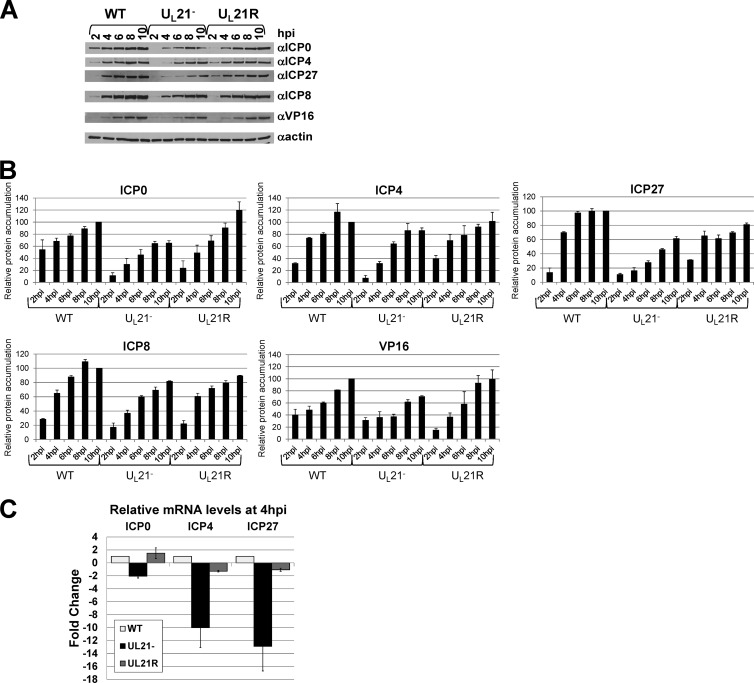
Fig 3.
Analysis of individual protein and mRNA levels. (A and B) Immunoblot analysis of steady-state ICP0, ICP4, ICP27, ICP8, VP16, and β-actin protein levels at early times in WT, UL21−, and UL21R infections. Lysates of Vero cells infected with the WT, UL21− and UL21R viruses for 2, 4, 6, 8, or 10 h were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with antibodies specific to ICP0 (Abcam), ICP4 (Rumbaugh-Goodwin Institute for Cancer Research), ICP27 (Virusys), ICP8 (16), VP16 (Santa Cruz Biotechnology), and β-actin (Santa Cruz Biotechnology). The experiment was performed three times. (A) Autoradiographic visualization of immunoblots. Similar results were obtained in three experiments. (B) Quantification of immunoblots. For each experiment, bands were normalized to the signal obtained from the WT 10-hpi band. Normalized values from each experiment were used to calculate relative means. Error bars represent one standard deviation. (C) Relative quantitative real-time RT-PCR analysis of ICP0, ICP4, and ICP27 mRNA levels in Vero cells infected with the WT, UL21−, and UL21R viruses for 4 h. Primer pair sequences were as follows: ICP0, 5′-CCTCTCCGCATCACCACAGAAGCC-3′ and 5′-CAGGTCTCGGTCGCAGGGAAACAC-3′; ICP4, 5′-CCGCCGTCGCAGCCGTATC-3′ and 5′-CCCGCCCTCCTCCGTCTCC-3′; ICP27, 5′-GGTGTCGGGGTCGGAGAGAAGATG-3′ and 5′-GGCGGTGCGTGTCTAGGATTTCG-3′; 18S rRNA, 5′-CCAGTAAGTGCGGGTCATAAGC-3′ and 5′-GCCTCACTAAACCATCCAATCGG-3′. The values are arithmetic means from two experiments, each performed in triplicate. Error bars represent one standard deviation.