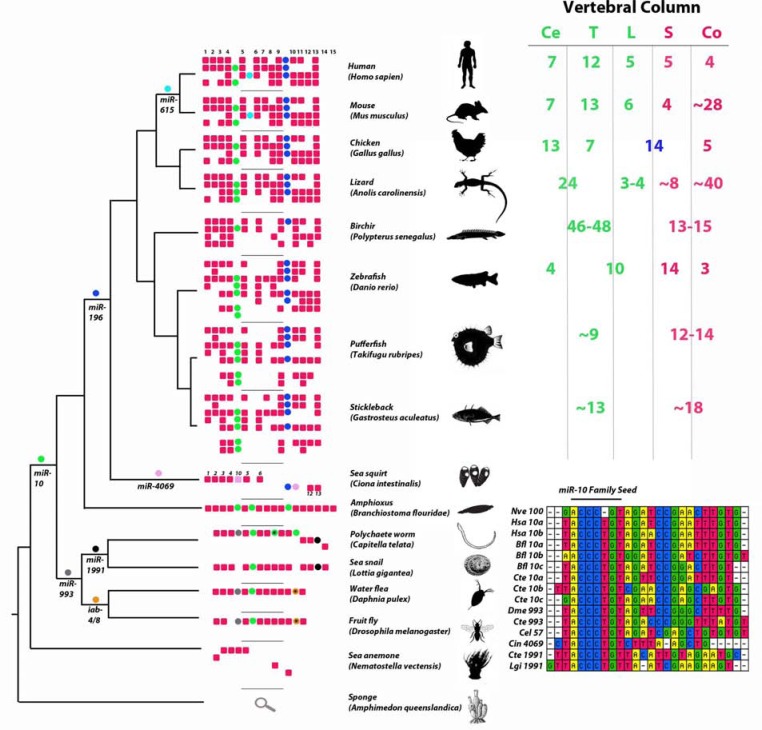
Fig. (1).
The Hox clusters among animals. Red boxes are Hox genes and colored circles are microRNA genes, miR-10 (green), miR-196 (dark blue), miR-615 (light blue), miR-993 (gray), miR-1991 (black), miR-4069 (pink), miR-1732 (brown) and iab-4/8 (orange). Genes clustered together in the genome are shown within the same row for a given Hox cluster. For example, human has four Hox clusters, amphioxus has a single Hox cluster and the sea anemone has seven Hox genes among four genomic scaffolds. Relationships among taxa are represented by the cladogram on the left. Variation in the number of vertebrae of the vertebral column is presented on the right. Ce (cervical), T (Thoracic), L (Lumbar), S (Sacral), and Co (Coccyx). Ce, T, and L are pre-caudal regions and colored green. S and Co are caudal regions and colored red. Note that neither Hox genes nor miRNA genes alone define the length or number of segments within the vertebral column. Alignment of miR-10 family Hox-embedded microRNAs (lower right). miR-100 from Nematostella vectensis is included in this aligment although it is not a Hox-embedded miRNA but a representative of the most deeply conserved member of the miR-10 family. Branchiostoma floridae (Bfl), Caenorhabtitis elegans (Cel), Ciona intestinalis (Cin), Capitella teleta (Cte), Drosophila melanogaster (Dme) Homo sapiens (Hsa), Lottia gigantea (Lgi), and Nematostella vectensis (Nve) [83-90].