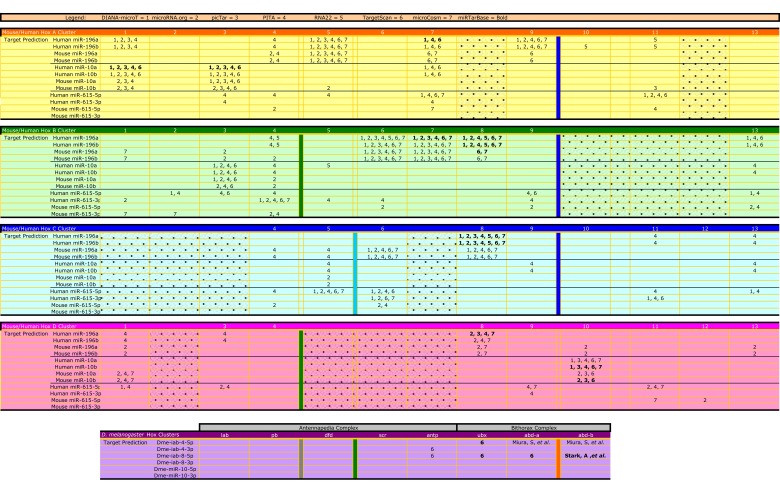
Fig. (3).
Summary of predicted Hox gene targets for miR-10, miR-196, miR-615 and miR-iab-4/8. Hox genes are predicted targets of Hox-associated miRNAs using several different prediction programs as shown here for the mouse, human, and fly. Each prediction program is given a numerical representative: DIANA-microT (1), microRNA.org (2), PicTar (3), PITA (4), RNA22 (5), TargetScan (6), and microcosm (7). These numbers are listed where they predict a given Hox:miR target interaction. For example, the HoxA cluster has 11 Hox genes and PITA (4) predicts Hoxa4 as a miR-196 target in humans and mouse. Shaded boxes denote genes that do not exist in that cluster (for example Hoxa8). The HoxA cluster is yellow, B cluster is green, C cluster is blue, D cluster is pink, and the D. melanogaster Hox complexes are purple. The positions of miRNA genes are marked by colored columns, green (miR-10), dark blue (miR-196), light blue (miR-615), orange (miR-iab 4/8), and gray (miR-993). Note that RNA22 was not queried for mouse targets and the fly predictions are directly from the published literature and/or TargetScan Fly. Although miR-993 is a Hox-associated miRNA in the fly, there are no known predictions for this miRNA. Targets with experimental support available in miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) are in bold. [62,88,91-98].