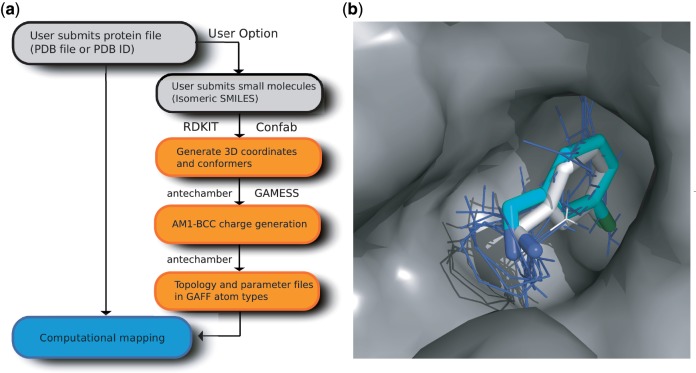
Figure 1.
Extended FTMAP server. (a) Schematic diagram of the updated implementation and the generation of force field parameters for user-supplied small molecules; programs used are indicated next to the appropriate boxes. (b) Mapping of an unbound structure of thrombin (PDB ID: 1HXF) using the small molecule C2A from a ligand-bound thrombin structure (PDB ID: 2C8Z) as a user-supplied additional probe. The lowest energy cluster of C2A (with the cluster shown as cyan sticks) overlaps the main consensus site (blue lines) from the mapping of the unbound thrombin structure using the standard probe set, and has an almost identical pose to the ligand from the bound structure (white sticks). Nitrogen and chlorine atoms are coloured blue and green, respectively.