Abstract
Circular permutation (CP) is a protein structural rearrangement phenomenon, through which nature allows structural homologs to have different locations of termini and thus varied activities, stabilities and functional properties. It can be applied in many fields of protein research and bioengineering. The limitation of applying CP lies in its technical complexity, high cost and uncertainty of the viability of the resulting protein variants. Not every position in a protein can be used to create a viable circular permutant, but there is still a lack of practical computational tools for evaluating the positional feasibility of CP before costly experiments are carried out. We have previously designed a comprehensive method for predicting viable CP cleavage sites in proteins. In this work, we implement that method into an efficient and user-friendly web server named CPred (CP site predictor), which is supposed to be helpful to promote fundamental researches and biotechnological applications of CP. The CPred is accessible at http://sarst.life.nthu.edu.tw/CPred.
INTRODUCTION
The protein structural rearrangement phenomenon termed circular permutation (CP) can be viewed as if the amino- and carboxyl-termini of a protein were relocated along the circularized sequence of the protein. Although the mechanisms underlying natural CP cases are not fully understood (1–5), many CPs have been observed in well-known protein families [see (6) for summaries of proposed mechanisms for CP and natural CP cases]. To study CP, many artificial circular permutants have been generated. The outcomes of these previous studies have indicated that as long as the CP site, i.e. the position for creating new termini, is not at a residue essential for protein folding or function, circular permutants usually retain native structures and biological functions (1,3,7–9), although the structural stabilities, folding mechanisms and enzymatic activities might be changed (10–15). These discoveries have made CP a new mutagenesis method for studying protein structure and function (16–18) and a bioengineering technique to modify the stability, solubility and activities of proteins (13,19–21). Moreover, the CP technique allows two proteins to be covalently linked at positions other than their native termini, facilitating the creation of several useful protein switches, molecular biosensors and fusion proteins (22–24). Despite these interesting applications, the implementation of CP is much more difficult, expensive and time-consuming compared with traditional mutagenesis. Most importantly, not all positions in a protein structure are permissible for CP. However, since practical software for predicting viable CP sites (i.e. positions leading to correctly folded and stable permutants) is still unavailable, researchers interested in CP-based protein engineering may rely on uneconomic trial-and-error for finding appropriate CP sites. To facilitate fundamental researches based on CP and biotech applications of the CP-based mutagenesis, we aim to develop an effective web-based tool for predicting viable CP site in this work.
CPs tend to occur at positions with high solvent accessibility (25), low sequence conservation and low ‘closeness’ (26), a structure-derived residue feature describing the amount of residues with which a given residue may interact directly or indirectly (27). However, predicting viable CP sites based on these properties yielded only marginal performance; the area under the receiver operating characteristic curve (AUC) values were all ≤0.7 (26). The major difficulty in developing CP viability predictors was the insufficiency of data, particularly the data of inviable CP sites (i.e. negative cases). In fact, the aforementioned predictors were developed and assessed with a data set composed of only one protein—dihydrofolate reductase (DHFR)—the entire 159-residued polypeptide of which had been subjected to systematic CPs (25). Although large data sets of CP-related protein structural homologs, such as the CP Database (CPDB) (28) and the database of GANGSTA+ Internet Services (GIS) (29), have been available since 2009, there is still a lack of negative cases. This is because most wet-lab researches only reported viable CP sites and bioinformatics CP-detecting methods could only identify existent (meaning viable) circular permutants. The DHFR data set contained only 73 negative cases, far from enough for developing reliable predictors.
Recently, we have established several highly different data sets for developing viable CP site prediction methods (30). Among them, nrCPDB-40 and nrGIS-40 contained thousands of proteins with machine-identified viable CP sites, whereas Data set T consisted of six proteins other than DHFR with both experimentally verified viable and inviable CP sites, expanding the number of negative cases by 2.4-fold (30). Based on these data sets, the sequence and structural preferences of CP were extensively examined (30). The identified preferences were quantified into numerous features to develop a CP viability prediction method that combined four machine learning algorithms: artificial neural networks, the support vector machine, a random forest and a hierarchical feature integration procedure (30). As trained with Data set T, this method achieved an AUC of 0.91 for the DHFR data set and a large-scale prediction sensitivity of ≥0.72 for either nrCPDB-40 or nrGIS-40. However, this effective CP site prediction method is not efficient. Due to heavy computational loads caused by several structural features and the time-consuming data flow through numerous prediction models, it took minutes to deal with one protein.
In our present work, we have implemented the developed CP viability prediction method into a user-friendly and quick response web server named CPred. Distributed computation techniques are used to accelerate the whole procedure, which now takes only seconds to make predictions. CPred is currently the most accurate method and is the only web server for predicting viable CP sites. We hope that it can be a good assistant for scientists and bioengineers to study and apply CP.
MATERIALS AND METHODS
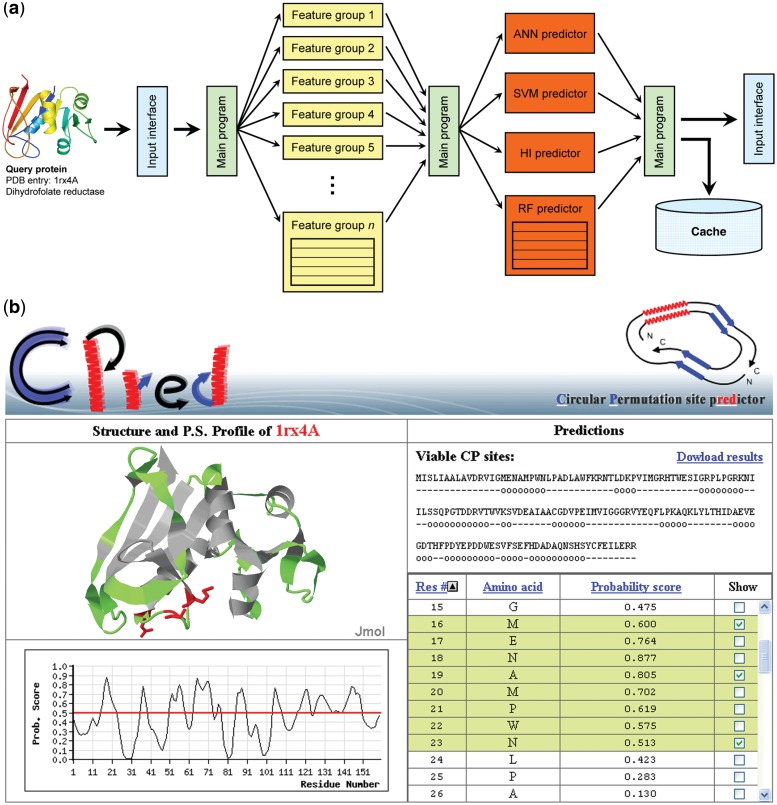
The flowchart of CPred is illustrated in Figure 1a. After receiving the query protein structure from the input module, the main program distributes to several processors the computation tasks of feature values, which are collected again by the main program. The main program then creates four threads running different machine learning predictors, the results of which are integrated and processed by the main program and are finally delivered to the output interface. If the protein structure is input by specifying a PDB [Protein Data Bank (31)] or a Structural Classification of Proteins (32) entry identifier, the calculated feature values and final results will be cached to ensure a quick response once the same protein is queried again in the future.
Figure 1.
The flowchart and output of CPred. (a) CPred is a viable circular permutation cleavage site prediction web server, which is working based on distributed computation techniques. After receiving the query protein data, the main program of CPred will extract feature values, execute machine learning subroutines, integrate the prediction results and deliver the final results to the output interface. The computation loads of many steps are distributed to several processors, as indicated by the radical arrow lines. Some structural features and machine learning methods require much more computation power than others; subroutines responsible for them, as represented by multicelled boxes, are designed by applying distributed techniques as well. (b) The output interface of CPred provides a list (lower right) and a graphic profile (lower left) of the probability scores of all residues in the input protein. The structure, along with predicted viable CP sites, is presented by an interactive Jmol (33) object (upper left), which allows the user to change the display mode (cartoon, spacefilled, etc.) and to rotate, resize and dissect the structure. A downloadable text version of the CPred results is provided as well (upper right). The structures shown in panel (a) and (b) were respectively rendered using PyMol (45) and Jmol (33).
Experimental data sets
Literature-derived data sets: Data set T and the DHFR data set
Information of inviable CP sites is rare, and it is extremely difficult to find a protein with both experimentally verified viable and inviable CP sites. Before our previous work (30), DHFR was the only data set for training and evaluating CP site predictors. By screening the literature, we had additionally collected six such proteins and established Data set T. Collectively, Data set T (76 viable and 100 inviable CP sites) and the DHFR data set (86 viable and 73 inviable CP sites) are the largest CP site data set currently available with both viable and inviable sites. Data set T and the DHFR data set shared very low sequence identities (<9%); the former was used to train and test our prediction system and the latter was used as an independent evaluation data set. These data sets are available in (30).
Database-extracted data sets: nrCPDB-40 and nrGIS-40
CPDB (28) and GIS (29) are the largest protein structural databases providing information about CP-related structural homologs. Previously, we had reduced these databases to 40% sequence identity non-redundant subsets, nrCPDB-40 and nrGIS-40 (30). Any protein in these two data sets that shared sequence identities >40% with any protein in the Data set T or DHFR were further eliminated. Finally, the nrCPDB-40 and nrGIS-40 data sets contained 1059 and 2814 proteins, respectively, and any two data sets among nrCPDB-40, nrGIS-40, Data set T and the DHFR data set shared <40% sequence identities [see (30) for details].
Non-redundant data set of CP site: nrCPsitecpdb-40
All CP sites of the proteins in nrCPDB-40 had been extracted (30). Each CP site was represented by a 20-residued segment. These 20-residued CP site representative segments were reduced to a 40% sequence identity non-redundant subset named nrCPsitecpdb-40 (1087 CP sites). Note that a protein may possess more than one viable CP site, and thus the number of non-redundant CP sites (1087) in the nrCPsitecpdb-40 data set is larger than the number of proteins (1059) in the nrCPDB-40 data set. The aforementioned data sets had been released as a part of the supporting data of (30).
Computation of feature values
The CPred system extracts 46 features from an input PDB file (see Supplementary Table S1). Based on a statistical significance test known as the permutation test (33), we had previously examined the sequence and secondary structural propensities of CP by comparing the compositions of single-, oligo- and coupled-residue patterns of amino acid sequence and several secondary structural strings between the CP site segments of nrCPsitecpdb-40 and the whole protein sequences of nrCPDB-40 (30). A secondary structural string, for instance, a Ramachandran structural string (34), is a text description of the secondary structure or backbone conformation of a protein. In CPred, these propensities are quantified by using the propensity scoring algorithm proposed in (30). Before CPred extracts tertiary structural features, the reduce program (35) is used to restore hydrogen atoms to the PDB file. Structure-derived residue measures and properties, e.g. the closeness (26), relative solvent accessibility, centroid distance measure (36), weighted contact number (37), farness (30) and the Gaussian Network Model-derived mean-square fluctuation (38–40), are then computed. All the obtained propensity scores and residue measures are standardized based on the conventional Z-score method (30) into real number features suitable for applying machine learning methods.
Application of machine learning methods
Four machine learning methods are applied in the CPred system: (i) a three-layered and back propagation-based (41,42) artificial neural network; (ii) a support vector machine established with the LIBSVM (43); (iii) a random forest of 500 decision trees generated by the C4.5 package (44); and (iv) a hierarchical feature integration procedure, in which features are hierarchically classified into a rooted tree that directs how the feature values are integrated into a single value (30). To efficiently integrate the prediction results from these methods, the output of every method for each residue has been designed to be a real number score between 0 and 1 [see (30) for algorithms]. Because of the range of value of these scores and their being conceptually in direct proportion to the chance that a case is a positive case, we have termed them ‘probability scores’ for convenience (30). Since these scores have the same range of values, to integrate the prediction power of various methods, we simply average their probability scores into an integrated score, based on which predictions are made. The feasibilities of these individual and integrated methods for predicting viable CP sites had been well established in (30), where the detailed algorithms, parameter settings and performance data are available. In the current work, a major problem in applying these methods lies in the fact that the data flow through the 500 decision trees of the random forest is very time-consuming. To solve this problem, as illustrated in Figure 1a, distributed computation techniques are used.
PERFORMANCE
Evaluations of the prediction system with cross-validation techniques and independent data sets
Before our work, the best viable CP site prediction method was developed based on the closeness measure, which achieved an AUC of 0.7 on the DHFR data set (26) and sensitivity values of 0.62 and 0.61 on the nrCPDB-40 and nrGIS-40 data sets, respectively (30). In our previous study (30), in which the core method of the current CPred system was developed, Data set T was used to establish the prediction model and the 10-fold cross-validated AUC, sensitivity, specificity and Matthews correlation coefficient values for this training data set were 0.91, 0.86, 0.79 and 0.63, respectively. Evaluating the established model with the independent data set DHFR, the aforementioned four performance measures were 0.91, 0.71, 0.92 and 0.64, respectively. A large-scale prediction test on this system registered sensitivity values 0.75 and 0.72 for nrCPDB-40 and nrGIS-40, respectively [refer to (30) for details]. These data indicated that the core method of CPred outperformed previous methods with little data set dependence or overfitting. Since the CPred server is running the same core programs, its performance measures assessed with these data sets are the same with the values listed earlier in the text. In the actual CPred web server, the prediction model is trained with a combined data set of Data set T and DHFR. Evaluations made based on this combined data set with 10-fold cross-validation and based on independent data sets nrCPDB-40 and nrGIS-40 show that accuracy of the actual server is improved as the amount of training data has increased (Table 1).
Table 1.
Performance of CP viability prediction of CPred
| Data set | Performance measure | Closeness | CPred |
|---|---|---|---|
| Training set (Data set T + DHFR data set)a | AUC | 0.753 | 0.940 |
| Sensitivity | 0.741 | 0.889 | |
| Specificity | 0.687 | 0.898 | |
| Matthews correlation coefficient | 0.428 | 0.787 | |
| nrCPDB-40 | Sensitivity | 0.622 | 0.746 |
| nrGIS-40 | Sensitivity | 0.614 | 0.719 |
aThese results were obtained with 10-fold cross-validation.
Evaluations of the developed probability score with information retrieval techniques
To help users interpret the results obtained with CPred, here we examine the average precisions of predictions at various decision thresholds of the probability score by performing 10-fold cross-validated information retrieval experiments. Table 2 demonstrates that a high threshold of probability score would retrieve fewer residues (i.e. a lower recall rate) but obtain a higher proportion of correct positive predictions (i.e. a higher precision) than a low threshold would. In the combined data set of Data set T and DHFR, any residue with a probability score ≥0.85 was an actual CP site (precision = 1). Since 82% of the residues predicted as viable CP sites (i.e. probability scores ≥0.5) were actual CP sites, this system is quite reliable. Experimenters expecting a high certainty about the viability of the created permutants may choose residues with probability scores ≥0.85 to apply CP; at this threshold, only 16% of all residues in a protein will be predicted as viable CP sites (i.e. the predicted positive fraction = 0.16).
Table 2.
Performance of CPred at various decision thresholds of the probability score
| Probability score | PPFa | Recall | Precision |
|---|---|---|---|
| ≥0.90 | 0.06 | 0.13 | 1.00 |
| ≥0.85 | 0.16 | 0.33 | 1.00 |
| ≥0.80 | 0.26 | 0.52 | 0.99 |
| ≥0.75 | 0.33 | 0.66 | 0.96 |
| ≥0.70 | 0.39 | 0.74 | 0.92 |
| ≥0.65 | 0.43 | 0.81 | 0.92 |
| ≥0.60 | 0.49 | 0.88 | 0.87 |
| ≥0.50 | 0.54 | 0.92 | 0.82 |
| ≥0.40 | 0.61 | 0.97 | 0.77 |
| ≥0.30 | 0.69 | 1.00 | 0.70 |
| ≥0.20 | 0.78 | 1.00 | 0.62 |
| ≥0.10 | 0.90 | 1.00 | 0.54 |
| ≥0.00 | 1.00 | 1.00 | 0.48 |
aPPF: predicted positive fraction, meaning the proportion of residues predicted as viable CP sites among all residues in the data set.
Speed evaluations
Since some structural features used by CPred such as the closeness (26) and the weighted contact number (37) have a high time complexity in computation and the random forest of CPred possesses many subpredictors, reducing the running time is an important task for developing a quick response server. By applying distributed computation techniques, the computation loads of several time-consuming steps are shared by many processors, greatly enhancing the efficiency of the server. As listed in Supplementary Figure S2, CPred takes only ∼3.4 s to make predictions for a protein with 150–200 residues; even for proteins as large as 600 residues, the average running time is <22 s. Without distributed computation, the running time for proteins with 150–200 and approximately 600 residues is, respectively, around 48.8 and 513.6 s. These assessments were performed on the actual CPred server machine, which is a Linux computer with two 3.33 GHz octa-core Intel Xeon CPUs and 128 GB RAM.
WEB SERVER DESCRIPTION
The query interface of CPred accepts three types of input, inclusive of a PDB entry, a Structural Classification of Proteins entry or a PDB file. After the user submits the query data, a notification page will appear to show the status of computation and provide an URL through which the results can be retrieved at a later time if the user decides not to wait. The outputs of CPred include a list of probability scores for all residues of the input protein and an interactive Jmol (33) graphical display of the protein structure that demonstrates the predicted CP sites (Figure 1b). The list of results can be reordered according to the residue number, amino acid type or the probability score.
APPLICATIONS AND FUTURE WORKS
CPred is a user-friendly web server for predicting possible cleavage sites for creating correctly folded and stable circular permutants of proteins. It provides a convenient probability score to help the user select suitable CP cutting sites. An interesting application of CP is to create fusion proteins with tethered sites different from the native termini (22–24). To our knowledge, for every CP-involved fusion protein that has been created, CP was introduced into just one of the fused polypeptides. This is perhaps because of the difficulty in generating two viable circular permutants at the same time. The convenient probability score of CPred may potentiate the production of fusion circular permutants. CP has long been applied to study protein folding. The ability of CPred to predict CP sites implies that it can be used in reverse to predict residues important to folding. Improving protein function is also a useful application of CP. Since residues with low probability scores are unlikely to form viable—not to mention functionally improved—permutants, CPred holds promise for bioengineering by screening out improbable cases. To improve CPred, additional data will be continually collected for training the prediction model. The current CPred server requires protein structures for making predictions. However, there are so many proteins without determined structures. We supposed that a sequence-based viable CP site predictor will further facilitate the application of CP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figure 2 and Supplementary References [34,46–50].
FUNDING
Funding for open access charge: The National Science Council, Taiwan [100-2745-B-009-001-ASP, Academic Summit Program of National Science Council to J.-K.H.]. National Science Council, Taiwan [100-2627-B-007-005 and 100-2319-B-400-001 to P.-C.L.]; The ‘Center for Bioinformatics Research of Aiming for the Top University Program’ of National Chiao Tung University and the Ministry of Education, Taiwan, R.O.C.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Prof. Chen-Hsiang Yeang at Academia Sinica, Taiwan, for insightful suggestions, and Linwei Technology Ltd. for securing the server machines. The authors of all algorithms and software used in the developed server are sincerely acknowledged.
REFERENCES
- 1.Cunningham BA, Hemperly JJ, Hopp TP, Edelman GM. Favin versus concanavalin A: circularly permuted amino acid sequences. Proc. Natl Acad. Sci. USA. 1979;76:3218–3222. doi: 10.1073/pnas.76.7.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ponting CP, Russell RB. Swaposins: circular permutations within genes encoding saposin homologues. Trends. Biochem. Sci. 1995;20:179–180. doi: 10.1016/s0968-0004(00)89003-9. [DOI] [PubMed] [Google Scholar]
- 3.Lindqvist Y, Schneider G. Circular permutations of natural protein sequences: structural evidence. Curr. Opin. Struct. Biol. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- 4.Uliel S, Fliess A, Unger R. Naturally occurring circular permutations in proteins. Protein. Eng. 2001;14:533–542. doi: 10.1093/protein/14.8.533. [DOI] [PubMed] [Google Scholar]
- 5.Weiner J, 3rd, Thomas G, Bornberg-Bauer E. Rapid motif-based prediction of circular permutations in multi-domain proteins. Bioinformatics. 2005;21:932–937. doi: 10.1093/bioinformatics/bti085. [DOI] [PubMed] [Google Scholar]
- 6.Lo WC, Lyu PC. CPSARST: an efficient circular permutation search tool applied to the detection of novel protein structural relationships. Genome. Biol. 2008;9:R11. doi: 10.1186/gb-2008-9-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribeiro EA, Jr, Ramos CH. Circular permutation and deletion studies of myoglobin indicate that the correct position of its N-terminus is required for native stability and solubility but not for native-like heme binding and folding. Biochemistry. 2005;44:4699–4709. doi: 10.1021/bi047908c. [DOI] [PubMed] [Google Scholar]
- 8.Tsai LC, Shyur LF, Lee SH, Lin SS, Yuan HS. Crystal structure of a natural circularly permuted jellyroll protein: 1,3-1,4-beta-D-glucanase from Fibrobacter succinogenes. J. Mol. Biol. 2003;330:607–620. doi: 10.1016/s0022-2836(03)00630-2. [DOI] [PubMed] [Google Scholar]
- 9.Vogel C, Morea V. Duplication, divergence and formation of novel protein topologies. Bioessays. 2006;28:973–978. doi: 10.1002/bies.20474. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Shakhnovich EI. Different circular permutations produced different folding nuclei in proteins: a computational study. J. Mol. Biol. 2001;306:121–132. doi: 10.1006/jmbi.2000.4375. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Wang J, Wang W. Transition states for folding of circular-permuted proteins. Proteins. 2004;57:153–171. doi: 10.1002/prot.20175. [DOI] [PubMed] [Google Scholar]
- 12.Bulaj G, Koehn RE, Goldenberg DP. Alteration of the disulfide-coupled folding pathway of BPTI by circular permutation. Protein Sci. 2004;13:1182–1196. doi: 10.1110/ps.03563704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian Z, Lutz S. Improving the catalytic activity of Candida antarctica lipase B by circular permutation. J. Am. Chem. Soc. 2005;127:13466–13467. doi: 10.1021/ja053932h. [DOI] [PubMed] [Google Scholar]
- 14.Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 15.Todd AE, Orengo CA, Thornton JM. Plasticity of enzyme active sites. Trends. Biochem. Sci. 2002;27:419–426. doi: 10.1016/s0968-0004(02)02158-8. [DOI] [PubMed] [Google Scholar]
- 16.Anand B, Verma SK, Prakash B. Structural stabilization of GTP-binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic. Acids. Res. 2006;34:2196–2205. doi: 10.1093/nar/gkl178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebhard LG, Risso VA, Santos J, Ferreyra RG, Noguera ME, Ermacora MR. Mapping the distribution of conformational information throughout a protein sequence. J. Mol. Biol. 2006;358:280–288. doi: 10.1016/j.jmb.2006.01.095. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura T, Iwakura M. Circular permutation analysis as a method for distinction of functional elements in the M20 loop of Escherichia coli dihydrofolate reductase. J. Biol. Chem. 1999;274:19041–19047. doi: 10.1074/jbc.274.27.19041. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz TU, Walczak R, Blobel G. Circular permutation as a tool to reduce surface entropy triggers crystallization of the signal recognition particle receptor beta subunit. Protein Sci. 2004;13:2814–2818. doi: 10.1110/ps.04917504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y, Lutz S. Circular permutation: a different way to engineer enzyme structure and function. Trends Biotechnol. 2011;29:18–25. doi: 10.1016/j.tibtech.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Arnold FH. Fancy footwork in the sequence space shuffle. Nat. Biotechnol. 2006;24:328–330. doi: 10.1038/nbt0306-328. [DOI] [PubMed] [Google Scholar]
- 22.Kojima M, Ayabe K, Ueda H. Importance of terminal residues on circularly permutated Escherichia coli alkaline phosphatase with high specific activity. J. Biosci. Bioeng. 2005;100:197–202. doi: 10.1263/jbb.100.197. [DOI] [PubMed] [Google Scholar]
- 23.Ostermeier M. Engineering allosteric protein switches by domain insertion. Protein. Eng. Des. Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
- 24.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl Acad. Sci. USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwakura M, Nakamura T, Yamane C, Maki K. Systematic circular permutation of an entire protein reveals essential folding elements. Nat. Struct. Biol. 2000;7:580–585. doi: 10.1038/76811. [DOI] [PubMed] [Google Scholar]
- 26.Paszkiewicz KH, Sternberg MJ, Lappe M. Prediction of viable circular permutants using a graph theoretic approach. Bioinformatics. 2006;22:1353–1358. doi: 10.1093/bioinformatics/btl095. [DOI] [PubMed] [Google Scholar]
- 27.Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S. Network analysis of protein structures identifies functional residues. J. Mol. Biol. 2004;344:1135–1146. doi: 10.1016/j.jmb.2004.10.055. [DOI] [PubMed] [Google Scholar]
- 28.Lo WC, Lee CC, Lee CY, Lyu PC. CPDB: a database of circular permutation in proteins. Nucleic Acids Res. 2009;37:D328–D332. doi: 10.1093/nar/gkn679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerler A, Knapp EW. GIS: a comprehensive source for protein structure similarities. Nucleic Acids Res. 2010;38:W46–W52. doi: 10.1093/nar/gkq314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo WC, Dai T, Liu YY, Wang LF, Hwang JK, Lyu PC. Deciphering the preference and predicting the viability of circular permutations in proteins. PLoS One. 2012;7:e31791. doi: 10.1371/journal.pone.0031791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic. Acids. Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandonia JM, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hesterberg T, Moore DS, Monaghan S, Clipson A, Epstein R. Introduction to the Practice of Statistics. 5th edn. New York, NY: W.H. Freeman and Company; 2005. pp. 14.11–14.70. [Google Scholar]
- 34.Lo WC, Huang PJ, Chang CH, Lyu PC. Protein structural similarity search by Ramachandran codes. BMC Bioinformatics. 2007;8:307. doi: 10.1186/1471-2105-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 36.Shih CH, Huang SW, Yen SC, Lai YL, Yu SH, Hwang JK. A simple way to compute protein dynamics without a mechanical model. Proteins. 2007;68:34–38. doi: 10.1002/prot.21430. [DOI] [PubMed] [Google Scholar]
- 37.Lin CP, Huang SW, Lai YL, Yen SC, Shih CH, Lu CH, Huang CC, Hwang JK. Deriving protein dynamical properties from weighted protein contact number. Proteins. 2008;72:929–935. doi: 10.1002/prot.21983. [DOI] [PubMed] [Google Scholar]
- 38.Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 39.Haliloglu T, Bahar I, Erman B. Gaussian dynamics of folded proteins. Phys. Rev. Lett. 1997;79:3090–3093. [Google Scholar]
- 40.Zheng W. A unification of the elastic network model and the Gaussian network model for optimal description of protein conformational motions and fluctuations. Biophys. J. 2008;94:3853–3857. doi: 10.1529/biophysj.107.125831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elarabaty M. New Approach for the Solution of Modern Aerospace Systems Using the Artificial-Intelligence. Monterey, CA: 1989. Aiaa Computers in Aerospace VII Conference. Pts 1 and 2, pp. 300–310. [Google Scholar]
- 42.Werbos PJ. The Roots of Backpropagation: from Ordered Derivatives to Neural Networks and Political Forecasting. New York, NY: Wiley-Interscience; 1994. [Google Scholar]
- 43.Lin CH, Liu JC, Ho CH. Proceedings of the Second International Conference on Information Security and Assurance. Washington, DC, USA: IEEE Computer Society; 2008. Anomaly detection using LibSVM training tools; pp. 166–171. [Google Scholar]
- 44.Quinlan JR. Proceedings of the Thirteenth National Conference on Artificial Intelligence and the Eighth Innovative Applications of Artificial Intelligence Conference. Vol. 1 and 2. Portland, Oregon: AAAI Press / The MIT Press; 1996. Bagging, boosting, and C4.5; pp. 725–730. [Google Scholar]
- 45.DeLano WL. DeLano Scientific. San Carlos, CA, USA: 2002. [Google Scholar]
- 46.Pommie C, Levadoux S, Sabatier R, Lefranc G, Lefranc MP. IMGT standardized criteria for statistical analysis of immunoglobulin V-REGION amino acid properties. J. Mol. Recognit. 2004;17:17–32. doi: 10.1002/jmr.647. [DOI] [PubMed] [Google Scholar]
- 47.Timberlake KC. Chemistry. 5th edn. New York, NY: Haper-Collins Publishers Inc.; 1992. [Google Scholar]
- 48.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 4th edn. New York, NY: W.H. Freeman and Company; 2004. pp. 75–115. [Google Scholar]
- 49.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 50.Yang JM, Tung CH. Protein structure database search and evolutionary classification. Nucleic Acids Res. 2006;34:3646–3659. doi: 10.1093/nar/gkl395. [DOI] [PMC free article] [PubMed] [Google Scholar]