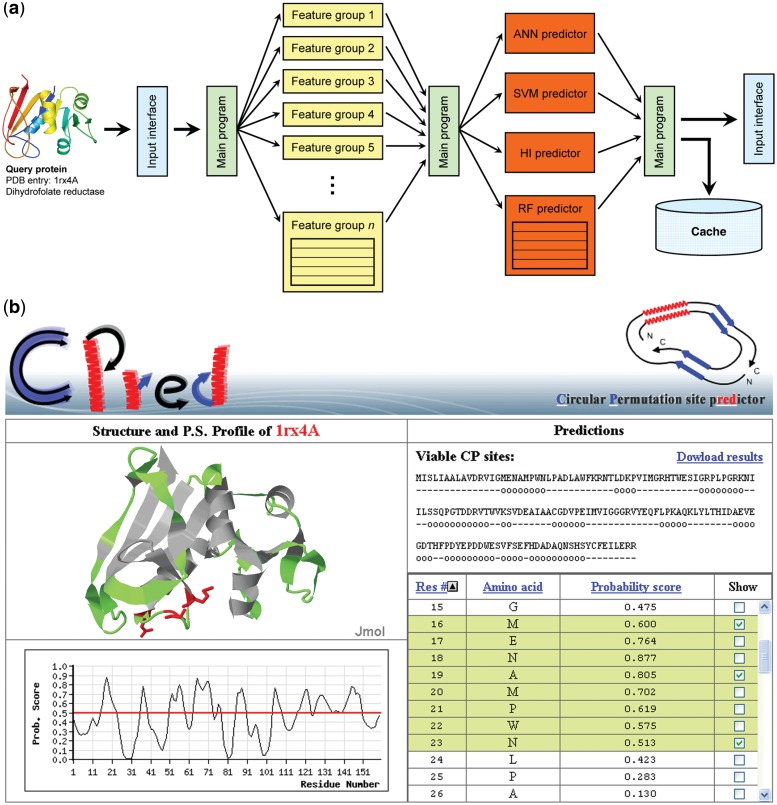
Figure 1.
The flowchart and output of CPred. (a) CPred is a viable circular permutation cleavage site prediction web server, which is working based on distributed computation techniques. After receiving the query protein data, the main program of CPred will extract feature values, execute machine learning subroutines, integrate the prediction results and deliver the final results to the output interface. The computation loads of many steps are distributed to several processors, as indicated by the radical arrow lines. Some structural features and machine learning methods require much more computation power than others; subroutines responsible for them, as represented by multicelled boxes, are designed by applying distributed techniques as well. (b) The output interface of CPred provides a list (lower right) and a graphic profile (lower left) of the probability scores of all residues in the input protein. The structure, along with predicted viable CP sites, is presented by an interactive Jmol (33) object (upper left), which allows the user to change the display mode (cartoon, spacefilled, etc.) and to rotate, resize and dissect the structure. A downloadable text version of the CPred results is provided as well (upper right). The structures shown in panel (a) and (b) were respectively rendered using PyMol (45) and Jmol (33).