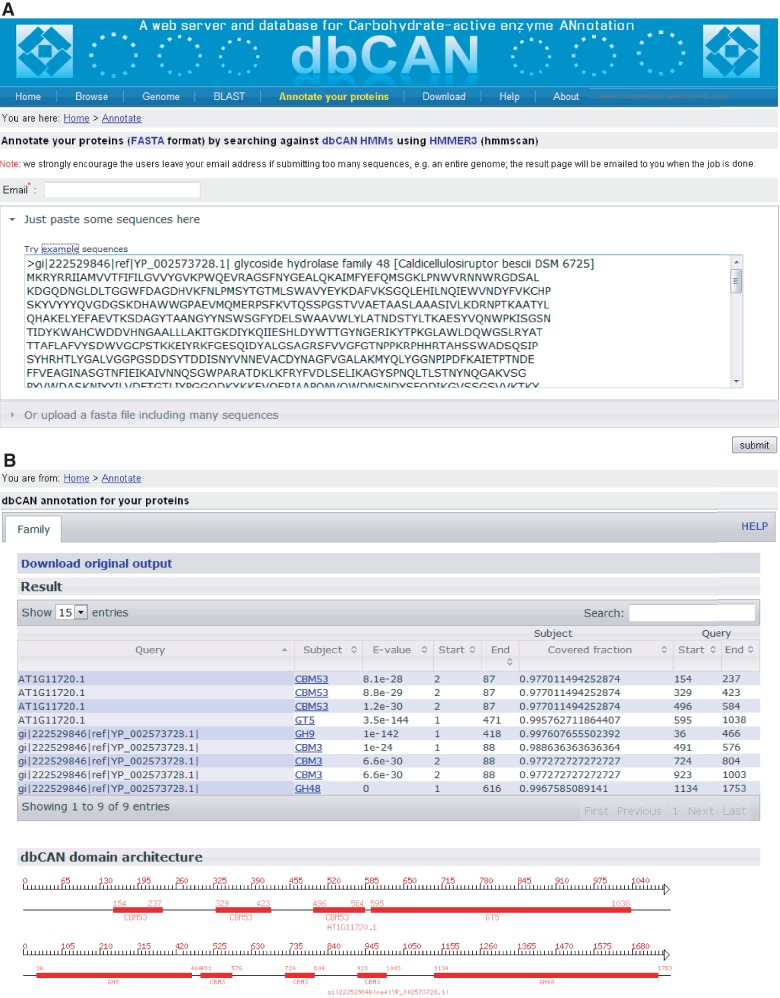
Figure 2.
Snapshots of dbCAN annotation server. (A) The query page, where users can paste some FASTA format protein sequences in the text box or upload a text file containing the FASTA sequences. Clicking on ‘submit’ will invoke the hmmscan program in the backend server to search the queried sequences against the dbCAN HMMs. (B) The result page, where users can download the raw output from the hmmscan run and view the processed tabular format output (if alignment length >80 amino acids, use E-value < 1e − 5, otherwise use E-value < 1e − 3). A diagram is shown in the bottom to illustrate the CAZyme domain architecture according to the positional information in the tabular output.