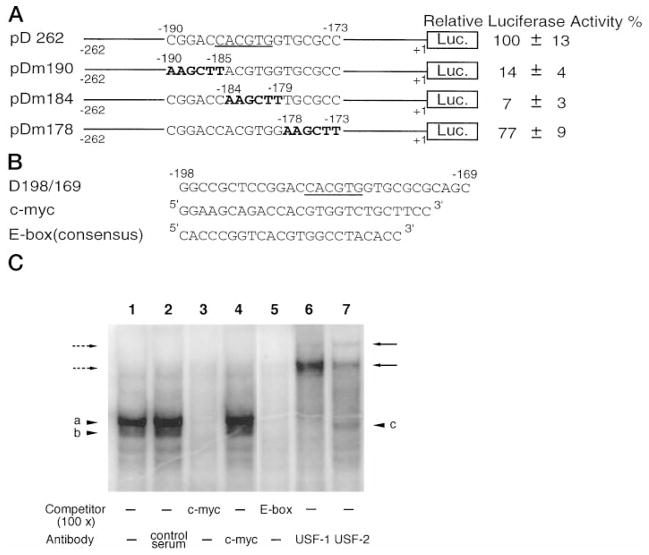
Fig. 5. Identification of the E box and its associated factors of the DOR promoter.
A, the effect of the E box mutation. The E box consensus in pD262 is underscored, and the mutated sequences in mutant constructs pDm190, pDm184, and pDm178 are shown in boldface. The activities of promoter constructs were examined by transient transfection assays in NS20Y cells. Luciferase activity was standardized to β-galactosidase activity and expressed as a percentage of the activity of the pD262 construct. The data on the right represent the mean values of relative luciferase activity (%) of pD262 from four independent transfection experiments. B, three oligonucleotides were used in EMSAs to identify the binding factors of the E box in the DOR promoter region. The oligonucleotide D198/169 contained the sequence from −198 to −169 of the DOR promoter. Both c-Myc and E box consensus oligonucleotides used as unlabeled competitors were commercial products from Santa Cruz Biotech-nology, Inc. C, EMSA was performed by using D198/169 as the probe with NS20Y nuclear extracts (lanes 1–7). Lane 1, control reaction; lane 2, 1 μl of control serum; lane 3, 100-fold excess of c-Myc competitor; lane 4, 1 μl of anti-c-Myc Ab; lane 5, 100-fold excess of E box competitor; lane 6, 1 μl of anti-USF1 Ab; lane 7, 1 μl of anti-USF2 Ab. The USF-D198/169 complexes are marked by arrowheads a and b. The putative USF1 homodimer is indicated by arrowhead c. The USF-D198/169 complexes recognized by anti-USF1 Ab are marked by broken arrows on the left. The USF-D198/169 complexes recognized by anti-USF2 Ab are marked by arrows on the right.