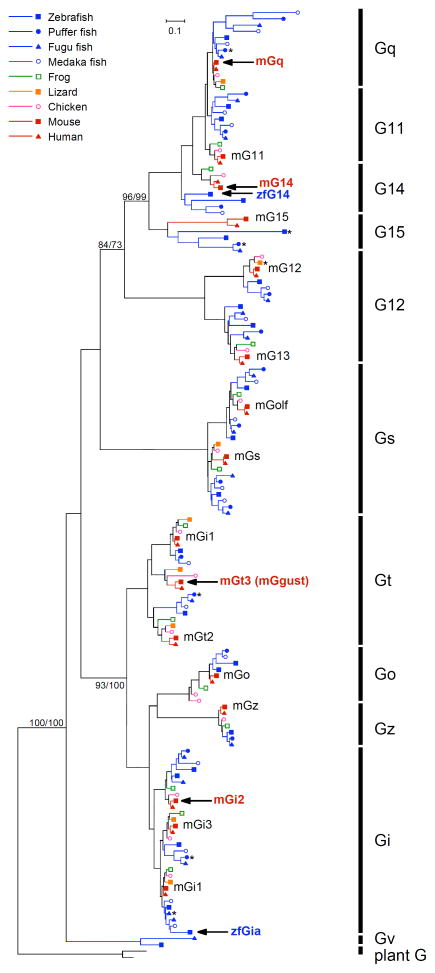
Fig. 1. Unrooted phylogenetic trees of G protein alpha subunits in vertebrates constructed by the maximum likelihood (ML) method.
Plant G alpha subunits are used as an outgroup. Bootstrap values over 70% calculated by the ML and neighbor-joining (NJ) method for branch support were presented in the order of ML/NJ for the major branch. Mouse and zebrafish G alpha genes expressed in taste receptor cells according to the present study and previous studies are shown in bold red and blue letters, respectively, with black arrows. Asterisks indicate genes that have been pseudogenized by insertion or deletion of a single nucleotide that was followed by a downstream in-frame stop codon.