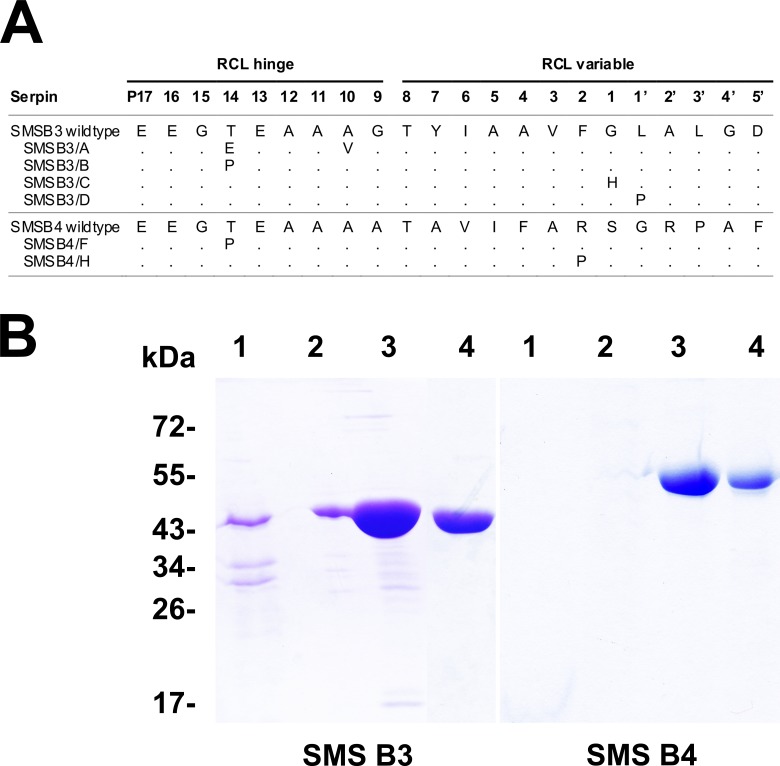
Figure 1. Sequence details and recombinant expression of SMSs and mutants. (A).
Shown are the sequence changes in the SMS mutants compared to wild type sequences. Numbering scheme according to [82]. Point mutations were introduced in the two segments of the RCL domain: i) the distal (P9-P5’) variable region that resembles the protease substrate and is attacked by a target protease, and ii) the proximal (P15-P9) hinge region that is well conserved and inserts into β-sheet A during inactivation of a protease in inhibitory serpins. (B) Purification and refolding of SMSB3 and SMSB4. Shown are immobilized nickel affinity chromatography fractions after separation by SDS-PAGE and Coomassie staining. Both SMSs revealed the expected bands of 47 kDa and 54 kDa, respectively. 1, loaded sample; 2, wash; 3, elution with 250 mM imidazole, 4, active serpin after refolding.