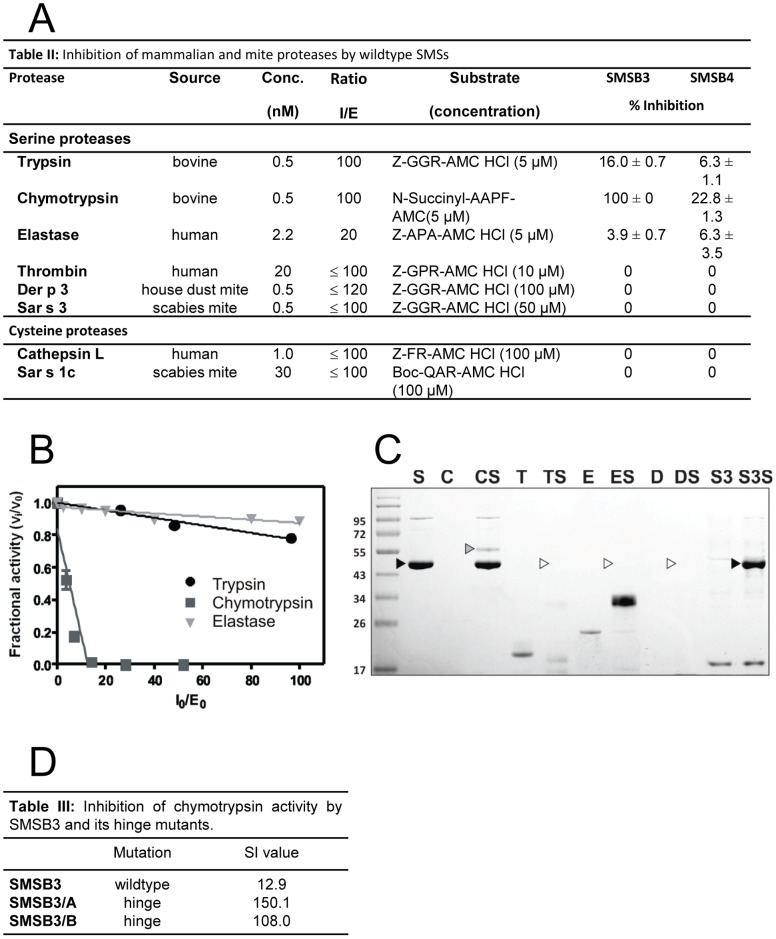
Figure 4. SMSs inhibit mammalian proteases and form serpin/protease complexes.
(A) Inhibitory profile of mammalian and mite proteases by wild type SMSs (overview). Proteases were pre-incubated with different concentrations of SMSB3 or SMSB4 at 37°C for 10 min in the appropriate reaction buffer. The reaction was started by addition of the corresponding fluorescent aminomethyl-coumarin substrate and enzyme inhibition was measured as change in fluorescence in comparison with controls. Shown are means ± SD (n = 3). (B) Stoichiometry of inhibition for SMSB3. The inhibitory activity was assessed by measuring residual enzyme activities of trypsin, chymotrypsin and leucocyte elastase. Enzymes were incubated with a 2-120:1 molar excess of SMSB3 at 37°C for 10 min in the appropriate reaction buffer. Residual enzyme activity was measured in triplicate (SD ≤5%) by adding the appropriate aminomethyl-coumarin peptide substrate and determining the reaction velocity as change in fluorescence. The fractional activity was the ratio of the velocity of inhibited enzyme (vi) over the velocity of uninhibited control (vo). The initial inhibitor/enzyme ratios ([I0]/[E0]) represent the molar excess of serpin over enzyme. (C) Complex formation between SMSB3 and serine proteases. Serpin/protease complexes were analyzed by SDS-PAGE under non-reducing conditions after pre-incubation of SMSB3 (black arrow) with each protease at 37°C for 15 min at serpin/protease ratios of 4∶1. Covalent complex formation was found with chymotrypsin (grey arrow), while SMSB3 was degraded by trypsin, elastase and Der p 3 (white arrows). C, chymotrypsin; D, house dust mite Der p 3; E, neutrophil elastase; S, SMSB3; S3, scabies mite Sar s 3; T, trypsin. (D) Effects of SMSB3 and two mutants on chymotrypsin activity.