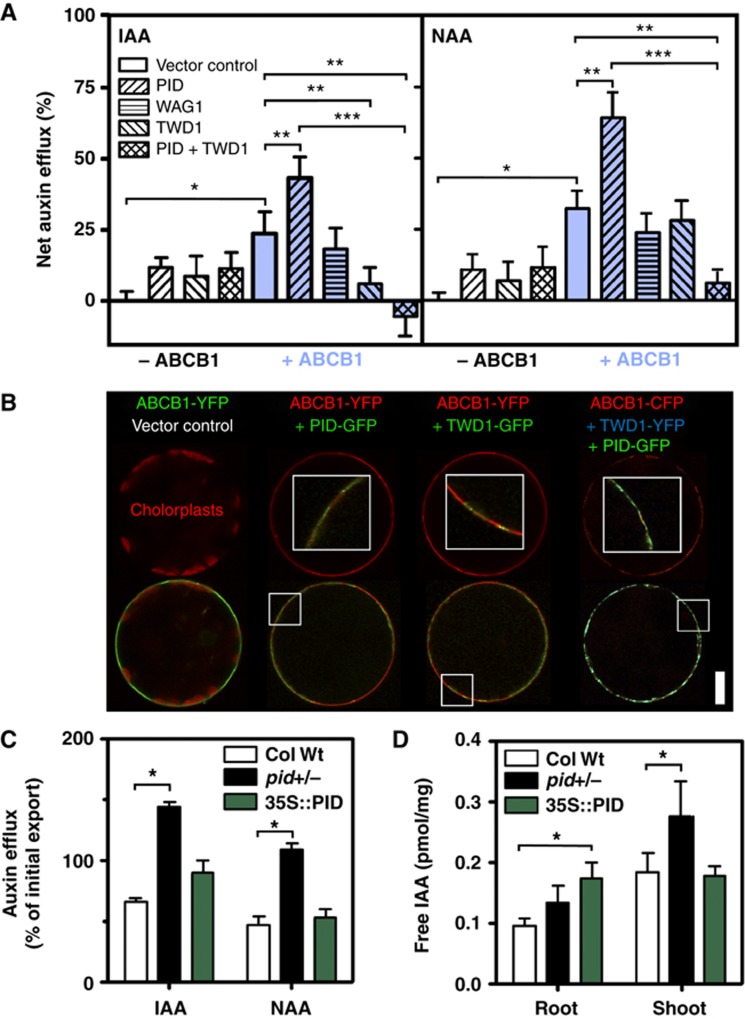
Figure 3.
PID negatively regulates ABCB1-mediated auxin efflux in planta. (A) Co-transfection of N. benthamiana protoplasts with PID specifically enhances ABCB1-mediated auxin (IAA and NAA) efflux in the absence of TWD1 but triple ABCB1/PID/TWD1 transfection strongly blocks ABCB1 activity (mean±s.e.; n=4). Significant differences (unpaired t-test with Welch’s correction, P<0.05) to vector control, ABCB1 and ABCB1/PID are indicated by one, two or three asterisks, respectively. (B) Co-transfection of PID (PID-GFP) and TWD1 (TWD1-GFP or TWD1-YFP) in N. benthamiana protoplasts does not significantly alter ABCB1 (ABCB1-YFP or ABCB1-CFP) location and expression in comparison to vector control co-expression (upper row). Note the colocalization of ABCB1, TWD1 and PID on the plasma membrane of tobacco protoplasts (lower row; insets in upper row show indicated details at higher magnification). For single and double co-expression controls, see Supplementary Figure S3H. Bar, 20 μm. (C) Efflux of native (IAA) and synthetic (NAA) auxin from Arabidopsis PID gain- (35S:PID) and loss-of-function (pid) protoplasts (means±s.e.; n=4; see Supplementary Figure S3C and D for time kinetics). (D) Free IAA levels determined by GC–MS are significantly elevated in the root of PID gain-of-function (35S:PID) and shoot of PID loss-of-function lines (pid+/−), respectively. Data are mean±s.e. (n=4 with each 40–50 seedlings). Note that material for C–D was prepared from heterozygous pid (pid+/−) plants since, due to technical limitations, a determination of homozygosity by shoot phenotyping or genotyping was not possible. Significant differences (unpaired t-test with Welch’s correction, P<0.05) between wild-type and mutant alleles/lines are indicated by asterisks.