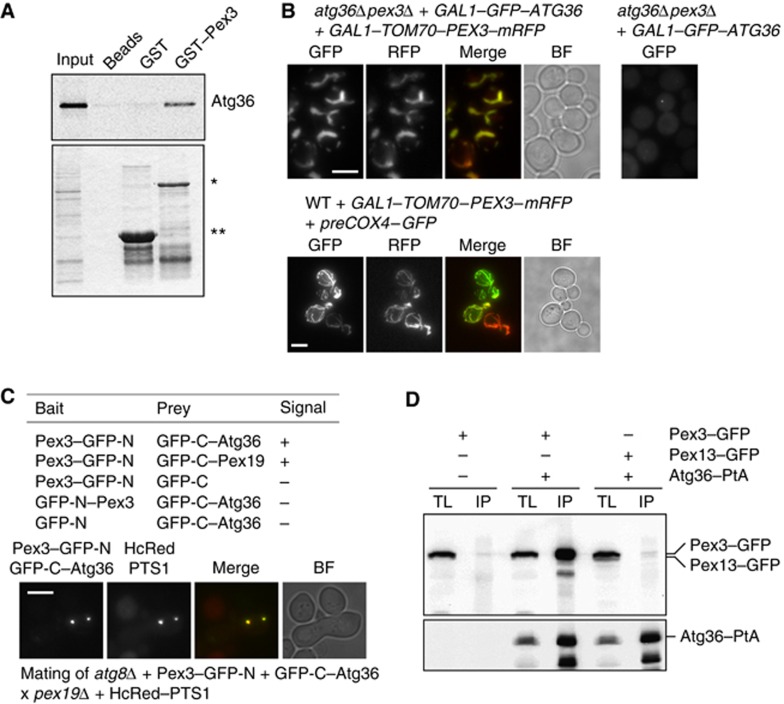
Figure 4.
Atg36 binds peroxisomes via Pex3. (A) An in vitro quick-coupled transcription/translation assay was performed using E. coli-produced GST–Pex3 (40–441) and GST bound to Glutathione Sepharose beads. After extensive washing of the bound protein, Atg36 synthesised in vitro in the presence of [35S]-Methionine was added. Following further washing, protein was eluted with GSH elution buffer and subjected to SDS–PAGE. Bound fractions and the Atg36 input were analysed by Coomassie staining (bottom panel) and PhosphoImaging (top panel). A beads-only sample was used as a further control. *Represents bound GST–Pex3 (40–441) and **represents bound GST. (B) atg36Δpex3Δ cells were transformed with plasmids encoding GAL1–GFP–Atg36 and a GAL1–Tom70–Pex3–mRFP chimeric protein (left panels, top) or GAL1–GFP–Atg36 (right panel, top). Expression was induced for 3 h. WT cells were transformed with GAL1–Tom70–Pex3–mRFP and preCox4–GFP (Motley et al, 2008) to confirm the mitochondrial localisation of Tom70–Pex3–mRFP. Bar, 5 μm. (C) Split-GFP analysis in atg8Δ cells. Cells expressing GFP halves were grown for 4 h in galactose medium, and scored for presence of fluorescence (−, no signal; +, fluorescent signal). Peroxisomes in cells expressing GFP–C-Atg36 plus Pex3–GFP-N were identified by mating with pex19Δ cells expressing HcRed–PTS1. Multiple fluorescent images were acquired in Z-axis and flattened into a single image. Bright-field image is a single plane. Bar, 5 μm. (D) Coimmunoprecipitation of Pex3 with Atg36. IP was performed in background strains of C13 abyss or C13–Atg36–PtA using plasmids expressing Pex3–GFP or Pex13–GFP under control of their endogenous promoters. Cells grown for 24 h to post-log phase in glucose medium. IgG Sepharose beads were used to immobilise Atg36–PtA from spheroplast yeast lysates. SDS–PAGE gels were probed with anti-GFP and PAP. Yeast lysates represent 5% of the lysate added to the beads and analysed by immunoblotting. TL, total lysate; IP, immunoprecipitate.