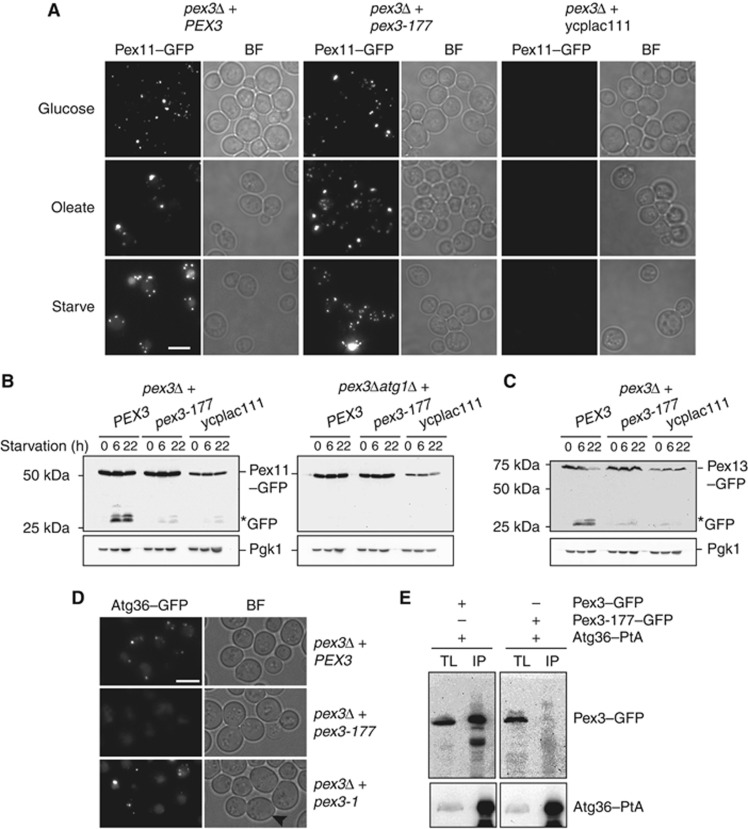
Figure 5.
The pex3-177 allele is affected in pexophagy. (A) Pex11–GFP pexophagy assayed by microscopy in pex3Δ cells and pex3Δatg1Δ cells transformed with plasmids containing either WT PEX3 or pex3-177, or with an empty plasmid ycplac111. Cells were grown in glucose, transferred to oleate medium for 18 h and switched to SD-N medium for 22 h. (B) Western blot analysis of same cells during starvation on SD-N medium. GFP* indicates the relative protease-resistant degradation product and reflects vacuolar breakdown. Time points for western blots indicated. (C) Pexophagy was assayed by Pex13–GFP breakdown in pex3Δ cells carrying plasmids as above. (D) Analysis of Atg36 localisation in pex3Δ cells expressing PEX3, pex3-177 and pex3-1. The mother cells of pex3-1 are devoid of peroxisomes (black arrowhead). (E) Coimmunoprecipitation of Pex3-177 with Atg36. IP was performed in the background strain of C13–Atg36–PtA using a plasmid expressing Pex3-177–GFP under control of its endogenous promoter. Cells grown for 24 h to post-log phase in glucose medium. IgG Sepharose beads were used to immobilise Atg36–PtA from spheroplast yeast lysates. SDS–PAGE gels were probed with anti-GFP and PAP. Yeast lysates represent 5% of the lysate added to the beads and analysed by immunoblotting. TL, total lysate; IP, immunoprecipitate. Comparison to Pex3–GFP IP (left panel).