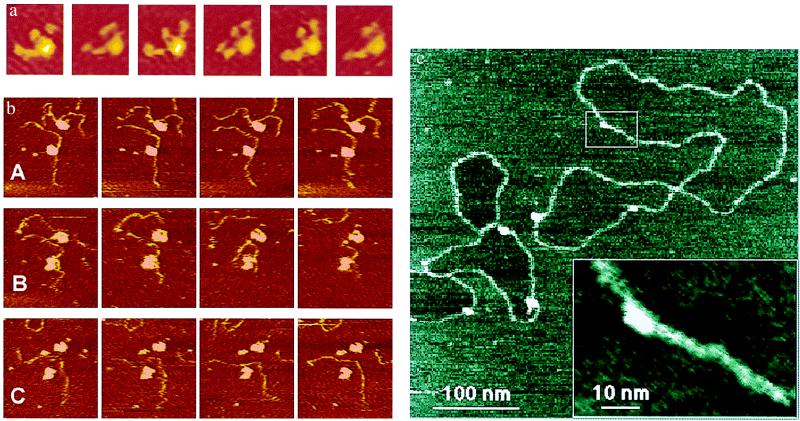
Figure 2.
(a) A laminin molecule moving its arms under fluid, taken by AFM. Six sequential frames from a 58-frame time-lapse movie show the arm movements of a molecule of Laminin-1 on mica. Images are 95 nm × 115 nm. Imaging buffer: 20 mM Mops/5 mM MgCl2/150 mM NaCl, pH 7.4. AFM cantilever was narrow V-shaped silicon nitride, 100-μm-long. See ref. 23. (b) Two active complexes of DNA with RNAP polymerase (RNAP) under fluid in an AFM. Escherichia coli RNAP transcription complexes were prepared with a 1,047-bp DNA template (3, 24). A, B, and C each show a series of four consecutive images at 42-s intervals. (A) DNA strands move near the surface in Zn(II) buffer. (B) These images were taken 3.5–6 min after the last image in A. RNAP transcribes and/or detaches from DNA strands after NTPs are introduced. (C) These images were taken 6–8 min after the last image in B. Zn(II) buffer is reintroduced. DNA images are 310 nm × 330 nm. (c) STM of humid plasmid DNA molecules. Hydration STM shows a high-resolution image of plasmid DNA, prepared on mica (an insulator) and imaged in humid air. The DNA shows a width of only 3 nm along much of its length. This image and other hydration STM images of DNA show an unusual feature: the mica surface inside some of the closed DNA loops appears to be slightly lower than the mica surface outside of closed DNA loops. The box overview marks the area shown in the Inset. This Inset is a cutout of a zoomed-in image taken immediately after the overview image. Imaging conditions: tunneling current, 0.5 pA; sample bias, −7 V; relative humidity, 65%. See ref. 15.