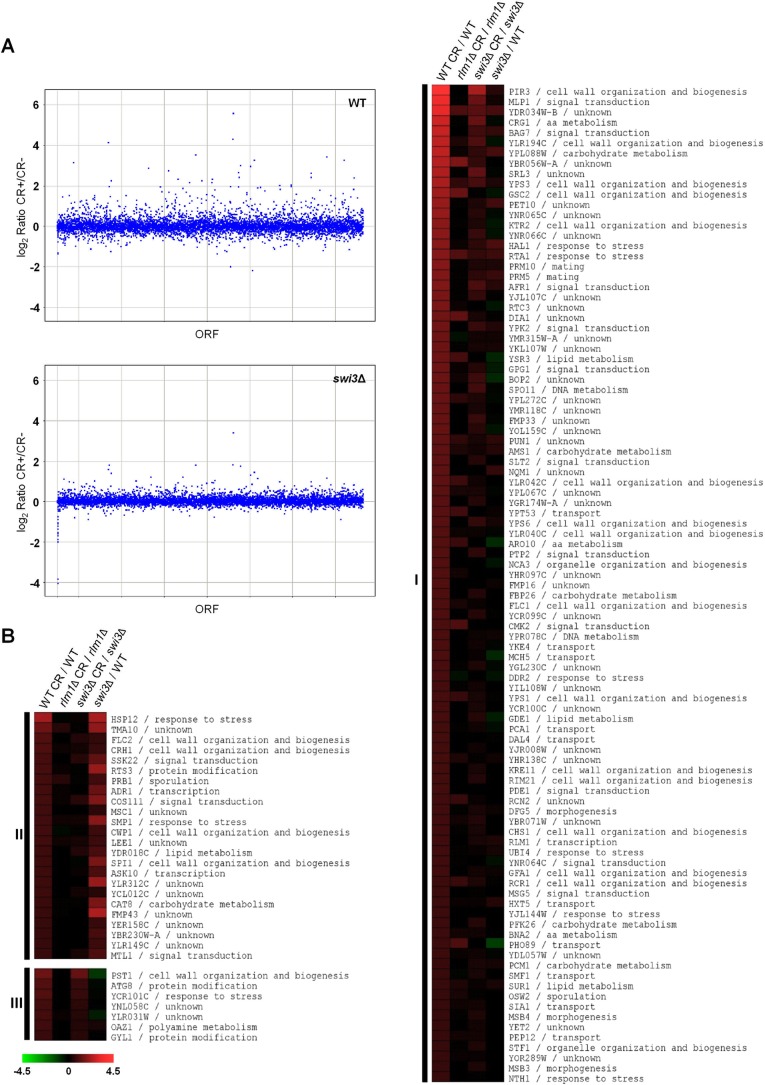
FIGURE 2:
Genome-wide expression profiles of WT (BY4741), swi3Δ, and rlm1Δ strains challenged with CR. (A) Scatter plot of normalized gene expression data in WT and swi3Δ strains. Gene expression ratios (treated/untreated) were plotted against their ORFs using Spotfire software. (B) Heat map obtained by MeV 4.6 software shows gene expression ratios comparing the transcriptional response to CR (treated vs. untreated) in the WT, rlm1Δ, and swi3Δ strains respectively (three columns on the left). Gene expression ratios of a swi3Δ mutant vs. a WT strain in the absence of stress are shown in the fourth column. The genes included in the analysis were those up-regulated in the WT strain by CR treatment. Genes were grouped together (clusters 1–3) on the basis of their dependence on Swi3 for basal and CR-mediated activation (see the text for details). Gene functional categories were assigned according to the information from the Biobase BioKnowledge Library (BKL; http://rous.mit.edu/index.php/Biobase_BioKnowledge_Library). The degree of color saturation represents the expression log2 ratio value, as indicated by the scale bar.