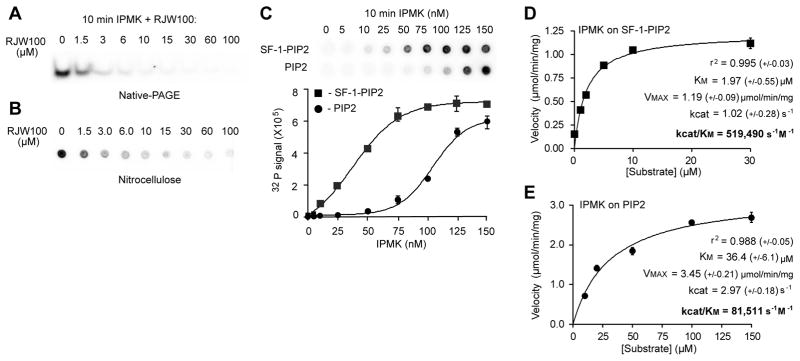
Fig. 2. SF-1/PIP2 Is A Bona Fide Substrate Of IPMK In Vitro.
(A) Autoradiography of IVK reactions of IPMK on SF-1/PIP2 substrate (1 μM) with increasing concentrations of RJW100 (SF-1 ligand, μM) analyzed by native-PAGE. (B) IPMK IVK assays on SF-1/PIP2 (1 μM) with increasing concentrations of RJW100 ligand added to displace PIP2, analyzed by nitrocellulose capture assay and quantified by phosphorimaging as described in Materials and Methods. (C) Comparison of IVK reactions using increasing amounts of IPMK and either 1 μM SF-1/PIP2 or PIP2-micelles, which were generated with phosphatidylserine carrier lipid as described in Methods. (D) IPMK reaction velocities plotted against increasing concentrations of SF-1/PIP2 or (E) PIP2-micelles. Data were fit to both non-linear (Michaelis-Menton) and linear double-reciprocal (Lineweaver-Burke) curves by GraphPad Prism software, refer to Fig S2 for double reciprocal plots. Kinetic parameters describing each enzyme/substrate pair, as indicated, (+/− represents standard error except r2, where +/− represents the absolute sum of squares for the curve fit).