Figure 2. Residual methylation in DKO1 is not caused by an inherent susceptibility to DNA methylation.
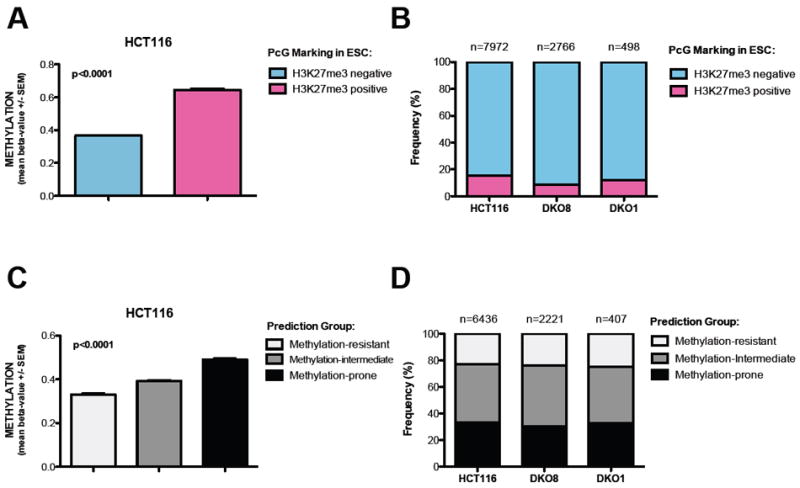
(A) Validation of H3K27me3 status in ESC as a predictive method for DNA methylation in HCT116 cells. Methylation status of ~27,000 CpG sites was determined by Infinium. T-test with Mann Whitney post-test. Data represent the mean ± SEM.
(B) Frequency of probes marked by H3K27me3 in ESC in the cohort of DNA methylated probes (Beta value >0.6) in HCT116, DKO8 and DKO1 cells.
(C) Validation of the predictive method based on genomic architecture (Estecio et al., 2010) in HCT116 cells. Methylation status of ~27,000 CpG sites was determined by Infinium. One way ANOVA with Kruskal-Wallis test. Data represent the mean ± SEM.
(D) Frequency of methylation-prone genes in the cohort of DNA methylated genes (Beta value >0.6) in HCT116, DKO8 and DKO1 cells.
