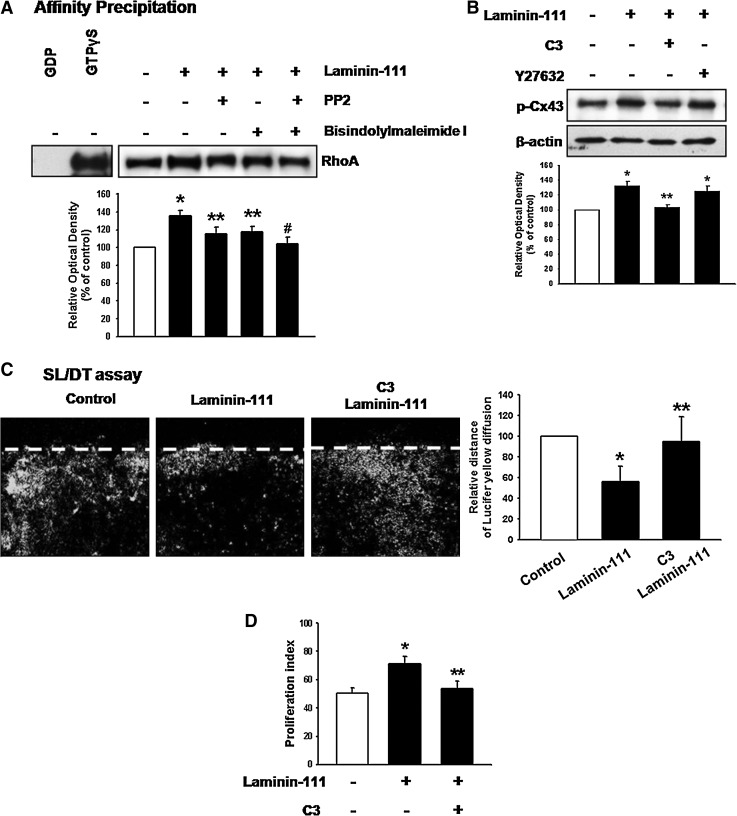
FIG. 3.
Involvement of Ras homolog gene family, member A (RhoA) on Cx43 phosphorylation. (A) Cells were treated with PP2 or/and bisindolylmaleimide I for 30 min before laminin-111 treatment and then loaded with GDP (negative control; lane 1) or GTPγS (positive control; lane 2) before affinity precipitation in the presence of 10 μg of glutathione-S-transferase- Rho Binding Domain on glutathione-sepharose beads. After each binding reaction at 4°C, the proteins bound to the beads were separated by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and examined for GTP-bound RhoA. The lower panel depicts the mean±SD of three independent experiments for each condition. *P<0.05 versus control; **P<0.05 versus laminin-111; #P<0.05 versus PP2 with laminin-111 or bisindolylmaleimide I with laminin-111. (B) Cells were treated with C3 (RhoA inhibitor; 1 μg/mL) or Y27632 (ROCK inhibitor; 10–6 M) for 30 min before laminin-111 treatment for 30 min, and the expression of p-Cx43 was detected by western blot. The lower panel depicts the mean±SD of three independent experiments for each condition as determined from densitometry relative to β-actin. *P<0.05 versus control; **P<0.05 versus laminin-111. (C) Cells were treated with C3 for 30 min before laminin-111 treatment for 30 min, and GJIC analysis was carried out using a scrape loading/dye transfer (SL/DT) assay, as described in the Materials and Methods section. The lower panel depicts the mean±SD of four independent experiments for each condition as determined from quantification of GJIC (the relative distance of Lucifer yellow diffusion compared with control). *P<0.05 versus control; **P<0.05 versus laminin-111. (D) Cells were treated with C3 for 30 min before laminin-111 treatment for 24 h, harvested, and subjected to propidium iodide (PI) staining for cell-cycle analysis by flow cytometry. Gates were configured manually to determine the percentage of cells in the S phase based on DNA content. Data are calculated using proliferation indices [(S+G2/M)/(G0/G1+S+G2/M)]×100 and reported as mean±SD of three independent experiments, each conducted in triplicate. *P<0.05 versus control;. **P<0.05 versus laminin-111. ROCK, Rho-associated kinase.