Abstract
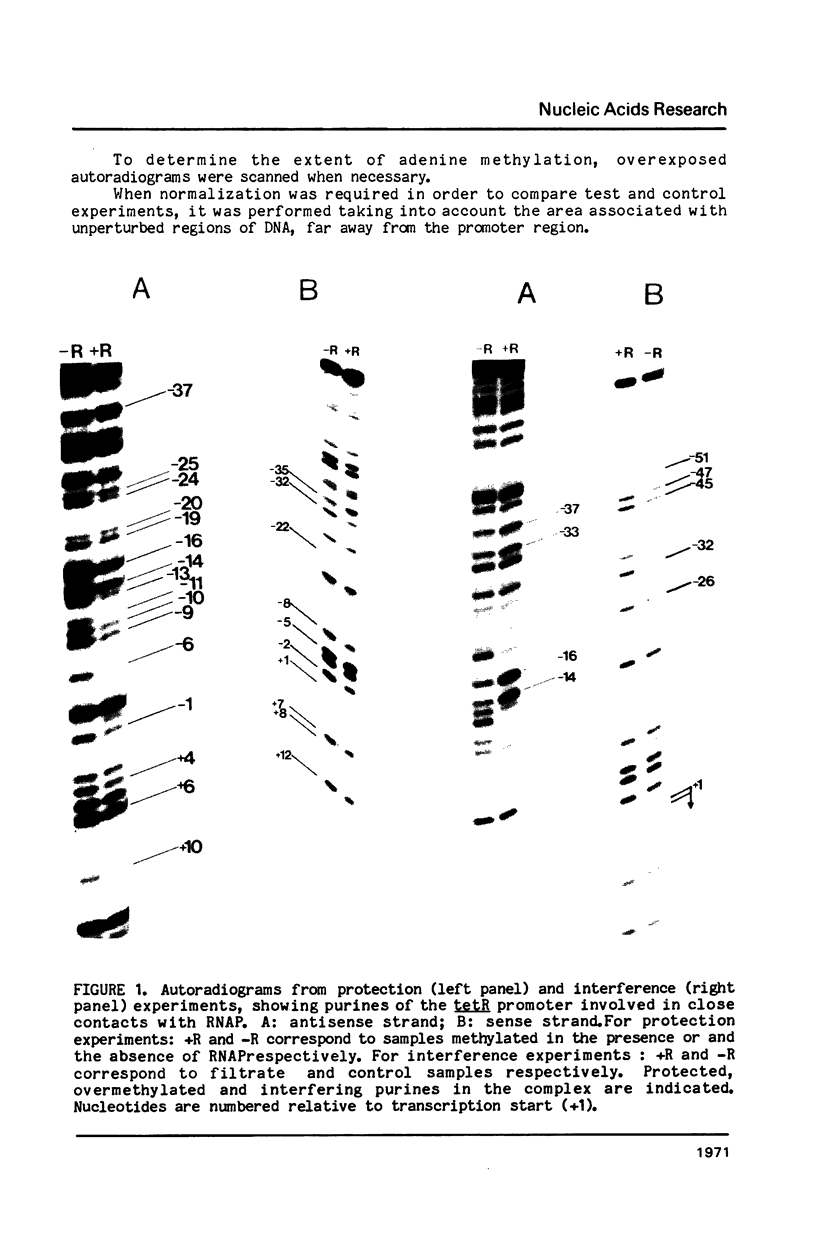
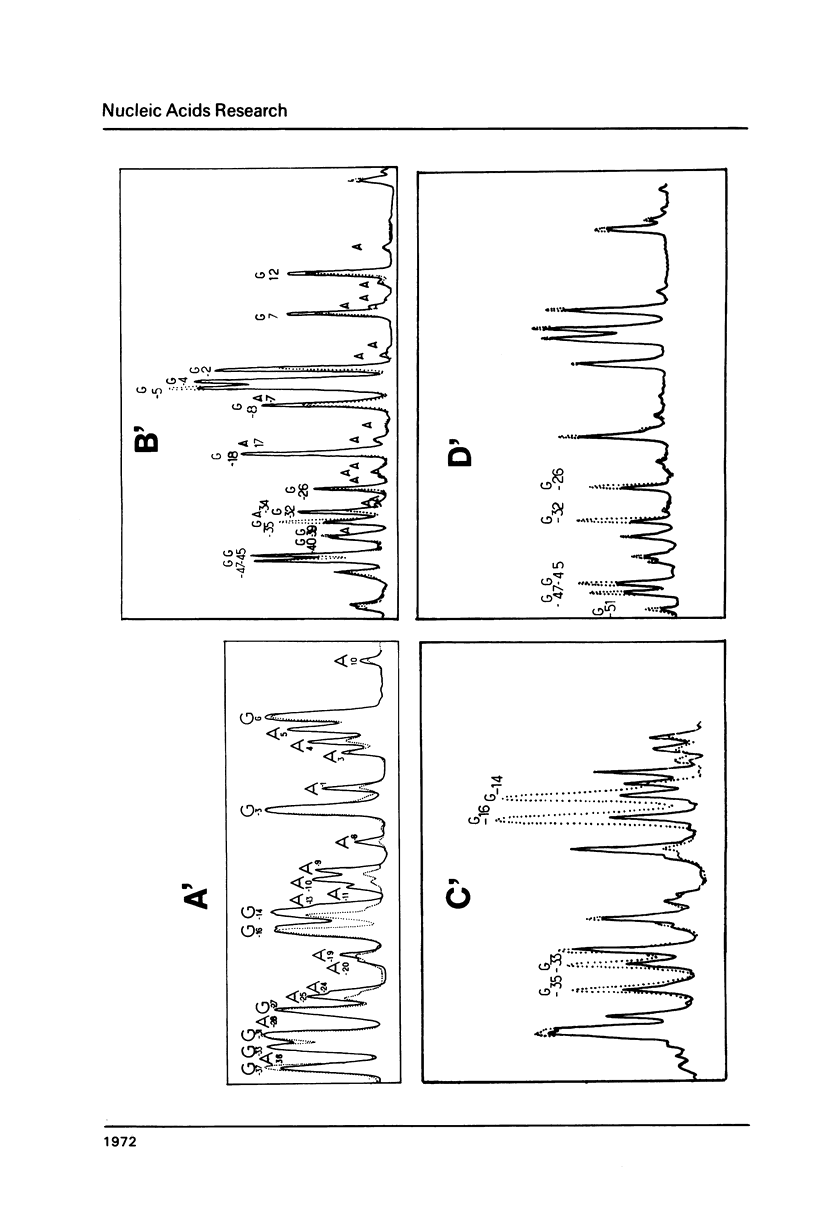
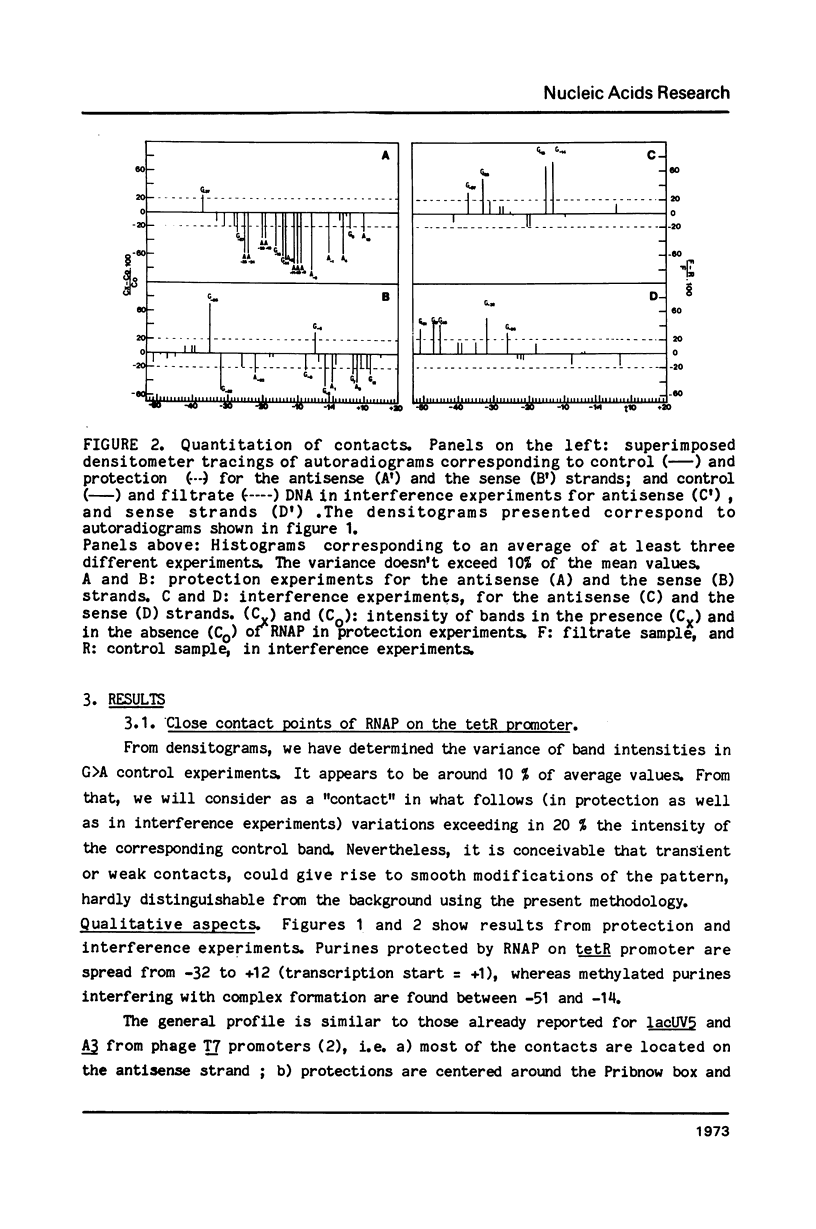
The interaction between E. coli RNA polymerase and the tetR promoter from pSC101, was studied by protection and premodification experiments, using dimethyl sulfate, methylation of single stranded cytosines, and DNAase I footprinting. Whereas qualitative and quantitative results from the chemical approach conform to patterns already displayed by other promoter systems, hypersensitive sites to DNAase I attack differ from those of other promoters. Distribution and nature of the contacts suggest that regions of the promoter sequence participates differently in complex formation. The involvement of major and minor grooves of the double helix in the complex with the enzyme, differs along the promoter. After a comparison of the results from seven different promoters, a pattern of conserved contacts seem to appear. Comparison of temperature dependence of local unwinding around the transcription start site (detected by the appearance of single stranded cytosines), and DNAase I footprinting, reveals that the process leading to stable complex formation can be achieved without disruption of base-pairing.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borowiec J. A., Gralla J. D. Supercoiling response of the lac ps promoter in vitro. J Mol Biol. 1985 Aug 20;184(4):587–598. doi: 10.1016/0022-2836(85)90305-5. [DOI] [PubMed] [Google Scholar]
- Buc H., McClure W. R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985 May 21;24(11):2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Busby S., Truelle N., Spassky A., Dreyfus M., Buc H. The selection and characterisation of two novel mutations in the overlapping promoters of the Escherichia coli galactose operon. Gene. 1984 May;28(2):201–209. doi: 10.1016/0378-1119(84)90257-9. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J., Nierman W. C., Wiggs J., Neff N. A quantitative assay for bacterial RNA polymerases. J Biol Chem. 1979 Oct 25;254(20):10061–10069. [PubMed] [Google Scholar]
- Chenchick A., Beabealashvilli R., Mirzabekov A. Topography of interaction of Escherichia coli RNA polymerase subunits with lac UV5 promoter. FEBS Lett. 1981 Jun 1;128(1):46–50. doi: 10.1016/0014-5793(81)81076-9. [DOI] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Ehrlich R., Larousse A., Jacquet M. A., Marin M., Reiss C. In vitro transcription initiation from three different Escherichia coli promoters. Effect of supercoiling. Eur J Biochem. 1985 Apr 15;148(2):293–298. doi: 10.1111/j.1432-1033.1985.tb08838.x. [DOI] [PubMed] [Google Scholar]
- Ehrlich R., Marin M., Larousse A., Gabarro-Arpa J., Schmitt B., Reiss C. Promoter recognition and transcription initiation in E. coli. Folia Biol (Praha) 1984;30(Spec No):105–118. [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen W., Schollmeier K., Gatz C. Control of expression of the Tn10-encoded tetracycline resistance operon. II. Interaction of RNA polymerase and TET repressor with the tet operon regulatory region. J Mol Biol. 1984 Jan 15;172(2):185–201. doi: 10.1016/s0022-2836(84)80037-6. [DOI] [PubMed] [Google Scholar]
- Hofer B., Müller D., Köster H. The pathway of E. coli RNA polymerase-promoter complex formation as visualized by footprinting. Nucleic Acids Res. 1985 Aug 26;13(16):5995–6013. doi: 10.1093/nar/13.16.5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Munson L. M., Mandecki W., Caruthers M. H., Reznikoff W. S. Oligonucleotide mutagenesis of the lacPUV5 promoter. Nucleic Acids Res. 1984 May 11;12(9):4011–4017. doi: 10.1093/nar/12.9.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. Promoter helical structure variation at the Escherichia coli polymerase interaction sites. J Biol Chem. 1984 Jun 10;259(11):6798–6805. [PubMed] [Google Scholar]
- Roe J. H., Burgess R. R., Record M. T., Jr Kinetics and mechanism of the interaction of Escherichia coli RNA polymerase with the lambda PR promoter. J Mol Biol. 1984 Jul 15;176(4):495–522. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- Roe J. H., Burgess R. R., Record M. T., Jr Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985 Aug 5;184(3):441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Spassky A., Kirkegaard K., Buc H. Changes in the DNA structure of the lac UV5 promoter during formation of an open complex with Escherichia coli RNA polymerase. Biochemistry. 1985 May 21;24(11):2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Sequence-dependent variations of B-DNA structure and protein-DNA recognition. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):271–278. doi: 10.1101/sqb.1983.047.01.032. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]