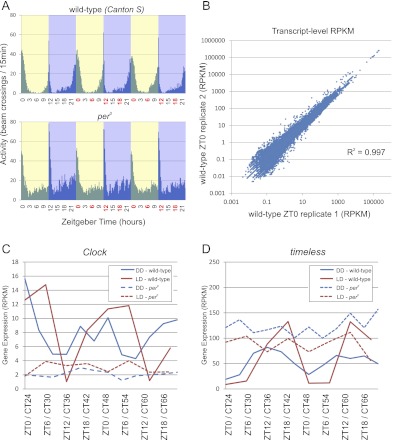
Figure 1.
RNA-seq accurately measures circadian transcriptional rhythms. (A) Locomotor activity rhythms were monitored using an automated infrared beam-break apparatus in wild-type and per0 flies entrained to a 12 h:12 h light:dark (LD) environment (N = 32 flies for each experiment). Plots are histograms of beam-breaks binned at a 20-min resolution. At the times indicated in red, brains were dissected and total RNA was purified for amplification and RNA-seq analysis. (B) Raw RNA-seq reads were aligned to the reference fly genome and transcriptome using the RUM read mapping algorithm, and expression levels (presented as reads per kilobase per million reads, RPKM) were calculated. Transcript expression levels exhibit a high degree of reproducibility between biological replicates with R-squared values on the order of 0.99. Expression profiles for Clock (C) and timeless (D) show consistent patterns of circadian oscillation in wild-type brains (solid lines) as measured by RNA-seq. Period-null mutation (dashed lines) disrupts the normal circadian rhythmicity of each of these transcripts. Clock and timeless oscillate in both LD (red) and DD (blue) with expected phases, and their amplitudes are damped in DD relative to LD, also as expected. The x-axis labels show the Zeitgeber time (ZT) for LD experiments and circadian time (CT) for wild-type Canton-S flies in DD.