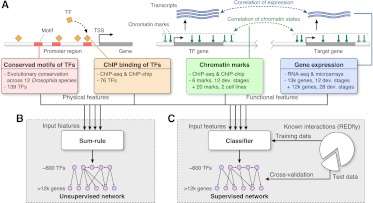
Figure 1.
Overview of inference methodology for supervised and unsupervised functional regulatory networks. (A) The four types of input features to the network inference algorithms are: evolutionary conserved motifs (red) and ChIP-based binding (orange) of TFs near the transcription start site (TSS) of target genes (physical features), and correlation of chromatin (green) and expression (blue) profiles between TFs and target genes (functional features). (B) Unsupervised network inference: An integrative network is formed by adding the evidence from each input feature with equal weight (sum-rule). (C) Supervised network inference: The input features are used in a classifier that predicts for every TF–target pair the presence or absence of a regulatory interaction. The classifier is trained on a literature-curated set of known interactions (REDfly).