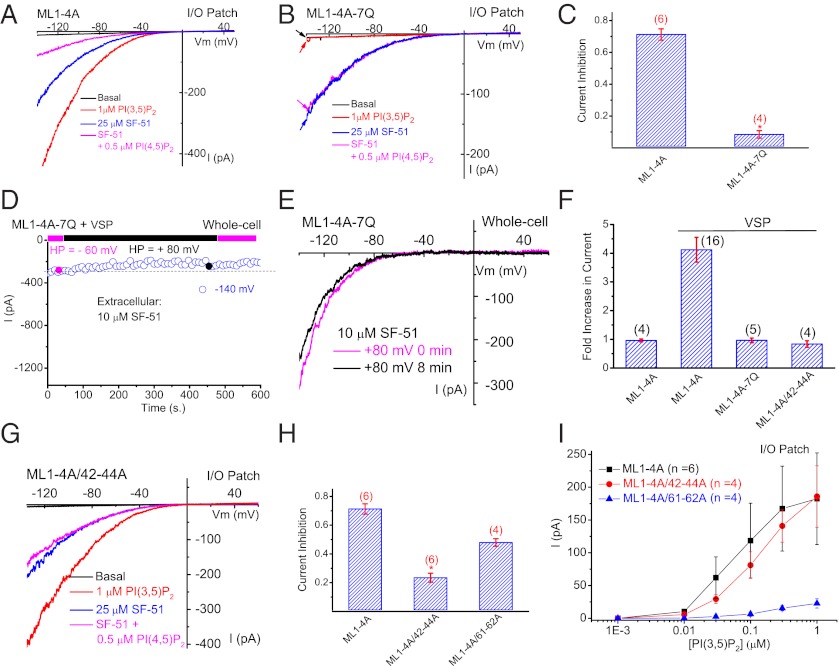
Fig. 4.
Structural determinants of PI(4,5)P2 inhibition of TRPML1. (A) In an I/O patch from ML1–4A–expressing HEK293 cells, IML1-4A activated by SF-51 (25 μM; bath application) was significantly inhibited by PI(4,5)P2 (0.5 μM). PI(3,5)P2 (1 μM) activated IML1-4A in the same patch. (B) SF-51–activated IML1-4A-7Q was insensitive to PI(4,5)P2 (0.5 μM), and IML1-4A-7Q was not activated by PI(3,5)P2 (1 μM) in the same patch. (C) Bar graph depicting the insensitivity of ML1-4A-7Q to PI(4,5)P2. (D and E) 7Q mutations abolished the effect of Ci-VSP activation (HP = +80 mV, 8 min) of ML1–4A. (F) 7Q mutations and triple mutations (R42A/R43A/R44A) abolished the stimulatory effect of Ci-VSP on ML1–4A. (G) ML1–4A/42–44A was only slightly inhibited by PI(4,5)P2, but robustly activated by PI(3,5)P2. (H) Bar graph depicting the sensitivity of ML1–4A/42–44A and ML1–4A/61–62A to PI(4,5)P2 inhibition (ML1–4A data were replotted from C for comparison). (I) ML1–4A/42–44A, but not ML1–4A/61–62A, remained sensitive to PI(3,5)P2 activation. Data are presented as mean ± SEM; numbers (n) are noted in parentheses. Statistical comparisons were performed using analysis of variance. *P < 0.05.