Abstract
Mass spectrometry-based serum metabolic profiling is a promising tool to analyse complex cancer associated metabolic alterations, which may broaden our pathophysiological understanding of the disease and may function as a source of new cancer-associated biomarkers. Highly standardized serum samples of patients suffering from colon cancer (n = 59) and controls (n = 58) were collected at the University Hospital Leipzig. We based our investigations on amino acid screening profiles using electrospray tandem-mass spectrometry. Metabolic profiles were evaluated using the Analyst 1.4.2 software. General, comparative and equivalence statistics were performed by R 2.12.2. 11 out of 26 serum amino acid concentrations were significantly different between colorectal cancer patients and healthy controls. We found a model including CEA, glycine, and tyrosine as best discriminating and superior to CEA alone with an AUROC of 0.878 (95% CI 0.815–0.941). Our serum metabolic profiling in colon cancer revealed multiple significant disease-associated alterations in the amino acid profile with promising diagnostic power. Further large-scale studies are necessary to elucidate the potential of our model also to discriminate between cancer and potential differential diagnoses. In conclusion, serum glycine and tyrosine in combination with CEA are superior to CEA for the discrimination between colorectal cancer patients and controls.
Keywords: Colorectal cancer, Mass spectrometry, Non-inferiority testing, Metabolic profiling
Introduction
Metabolomics, which is a sub-field of the new “omics” technologies, allows the systemic study of small-molecular weight metabolites expressed in a biological system (Ceglarek et al. 2009). These metabolites belong to diverse chemical classes such as amino acids, organic acids, fatty acids, or sugars (Chan et al. 2009; Dettmer et al. 2007). They are the final down-stream products of transcription and translation and thus are closest to the phenotype (Mamas et al. 2011). Hence, metabolomics promise to serve an important role in bridging the genotype-phenotype gap (Cascante and Marin 2008). In view of cancer disease previous research has particularly focused on the understanding of transcriptional regulation of cancer-associated gene expression, whereas less effort has been directed at investigations of metabolic alterations (Cardoso et al. 2007). Meanwhile, new mass spectrometry-based techniques allow a simultaneous and quantitative in-depth analysis of different metabolomic profiles in various biological samples (Ceglarek et al. 2009). These new approaches promise to enlighten the complex tumorigenesis associated metabolic alterations, which may accelerate the discovery of new diagnostic, prognostic, and predictive biomarkers. Despite this analytical evolution, however, profound knowledge and understanding of cancer metabolism is still lacking. This holds especially true for colorectal cancer (CRC), which is the third most common cause of cancer mortality in developed countries with an annual mortality of more than 500,000 cases (Herszenyi and Tulassay 2011). Colorectal cancer is among the top ten causes of death in Germany with an attributable fraction of 2% of all fatalities (DESTATIS, Statistisches Bundesamt Deutschland 2009). Comprising 16% of all cancer cases it is the second most common cancer type, and with 12–14% also the second most common cause of cancer death. Colonoscopy/sigmoidoscopy is still the “gold standard” in detecting colorectal carcinoma and high-risk adenomas, but its invasiveness, the experience of discomfort, the potential risks of complications, and the resources needed for the screening itself, when compared to FOBT, are disadvantages of concern (Bretthauer 2010). In contrast, the recommended iFOBT delivers sensitivity rates of 61–91% (Duffy et al. 2011), which are far from being satisfactory. Currently, there are no serum screening markers for CRC available. Carcinoembryonic antigen (CEA), the tumor marker of choice for CRC, is well suited for therapy monitoring, but also lacks the sensitivity needed for screening purposes (Shimwell et al. 2010). Earlier diagnosis of this cancer and early relapse monitoring after initial therapy are probably the best available options to improve patient survival. Established serum tumor markers such as CEA are useful to monitor the course of disease on and off treatment, but they lack the sensitivity and specificity criteria for screening stratification purposes (Tanaka et al. 2010). In this regard, mass spectrometry-based metabolic profiling may be valuable for the identification of new “disease signatures” and cancer-associated biomarkers. Preceding metabolic investigations primarily aimed at the discovery of metabolites significantly different between controls and CRC patients in serum, urine, and tissue (Wang et al. 2009) by applying standard test statistics [e.g. Student’s t-test, Wilcoxon-test, or the PCA (Principal Component Analysis)-based (O)PLS-DA (Ma et al. 2010; Qiu et al. 2009)] and revealed several potential marker metabolites. These preliminary results encouraged us to investigate the serum amino acid profiles and their alterations in CRC using a tandem mass-spectrometric approach (Fiedler et al. 2004; Mueller et al. 2003). In our study, we additionally considered two further aspects: to avoid pre-analytical flaws (Issaq et al. 2011) we strictly adhered to previously published protocols (Baumann et al. 2005; Brauer et al. 2011) as well as standardized sample processing (Ceglarek et al. 2002) and reporting (Fiehn et al. 2007). Second, to provide not only significant differences, but—as a core task of laboratory medicine—also statistically sound statements on their diagnostic surplus value, we additionally evaluated the markers we found and the conventional tumor marker CEA with respect to non-inferiority and superiority.
Materials and methods
Ethics statement
The study was approved by the ethics committee of the Medical Faculty of the University of Leipzig [Reg. No. 013-2005] and fulfills the requirements of the Helsinki declaration. All subjects gave written informed consent to participate in the study.
Patients and samples
Patients with CRC (n = 59) and respective controls (n = 58) were recruited at the University Hospital Leipzig in the context of a previously published study (Fiedler et al. 2009). Subjects were matched according to age and gender. Fasting blood sampling from patients was performed before initiation of specific therapy. Healthy controls called in for checkup showed no evidence of actual disease proven by physical examination and routine laboratory testing (differentials, C-reactive protein (CRP), creatinine, transaminases, alkaline phosphatase, γ-glutamyl transferase, bilirubin, tumor marker CEA). Venous serum samples were collected and stored by standardized techniques and protocols (Baumann et al. 2005; Brauer et al. 2011), including puncture of the cubital vein, 30–60 min coagulation at room temperature, centrifugation for 10 min at 1,400 g, immediate aliquotation and storage at –80°C until analysis.
Chemicals, standards and consumables
Materials
Methanol and isopropanol (gradient grade) were purchased from Merck (Darmstadt, Germany). The amino acid (AA) isotopes labelled standard kits (NSK-A, NSK-B, Cambridge Isotope Laboratories, Andover, USA) were used as internal standard. Water (HPLC grade) was obtained from J. T. Baker (Deventer, Netherlands). The derivatization reagent 3n butanolic HCl was made in-house by mixing 4:1 v/v of 1-butanol (for spectroscopy) from Merck (Darmstadt, Germany) and acetyl chloride (p.a.) from Sigma-Aldrich (Steinheim, Germany). 96-well polypropylene microtiter plates were purchased from Greiner Bio-One (Frickenhausen, Germany). Multifly needle sets and polypropylene serum monovettes with clotting activators were also obtained from Sarstedt. For sample storage 450 μl CryoTubes™ were purchased from Sarstedt (Nümbrecht, Germany).
Sample pretreatment
A sample derivatization protocol was used according to our formerly described procedures (Brauer et al. 2011; Ceglarek et al. 2002) to enhance the sensitivity of the mass spectrometric detection and thereby being able to minimize the sample volume (Harder et al. 2011). Serum samples were diluted 1:10 with methanol for protein precipitation. After centrifugation we placed 10 μl of the supernatant into 96 well polypropylene microtiter plates and diluted it with 100 μl of the internal standard solution. After evaporation at 70°C for 40 min, we added 60 μl of 3n butanolic-HCL for derivatization at 65°C for 18 min. Again, the residual solution was evaporated at 70°C for 40 min and then reconstituted with 150 μl of the mobile phase (1/1 v/v isopropanol/water). After 15 min of gentle shaking of the microtiter plate at room temperature, we analyzed the samples by flow injection analysis (FIA)-MS/MS. We aligned the samples in alternating series of 20 controls and cases on two microtiter plates and measured them in one analytical run on one day.
CEA
CEA was measured in serum samples by an electrochemiluminescence immunoassay (Roche, Germany) on Modular analytics E 170 analyzer (Roche, Germany) according to the manufacturer’s instructions.
Tandem mass spectrometry
An API 3000 tandem mass spectrometer (Applied Biosystems, Germany) using a Turbo Ion Spray Source (TIS) in combination with a HTC Pal autosampler and a PE 200 microgradient pump was used for flow injection analysis (FIA). 25 μl of the sample were directly injected at a flow rate of 80 μl/min in an analysis time of 1.5 min. We detected amino acids by a neutral loss scan of 102 in the mass range of 130–280 or multiple reaction monitoring (MRM). Quantitative analysis using internal standards was performed for 26 amino acids using ChemoView™ 1.4.2 (Applied Biosystems, Darmstadt, Germany). A comprehensive overview of mass transitions, internal standards, and performance data for the different amino acids can be found in Brauer et al. (2011).
Statistical analysis
Statistical testing was performed using ‘R’ (R Development Core Team 2008) with the packages ‘nortest’ (Gross 2006), ‘pROC’ (Robin et al. 2010, 2011), ‘BMA’ (Raftery et al. 2010), ‘car’ (Fox and Weisberg 2011), ‘care’ (Zuber and Strimmer 2010a), ‘ltm’ (Rizopoulos 2010), ‘boot’ (Davison and Hinkley 1997), ‘stats’, and ‘rattle’ (Graham 2009). To test for normality we applied the Anderson–Darling test from ‘R’s ‘nortest’ package (Stephens 2006). Gender distribution in the groups was evaluated applying Fisher’s exact test, group-related differences for the remaining parameters by Wilcoxon’s test (both from R’s ‘stats’-package). For bootstrapping, we used ‘R’s ‘sample’ function as well as the ‘boot’ package [B = 999 runs with replacement, cf. Carpenter and Bithell (2000)] to compute robust estimates for the 95% confidence intervals (CIs) of the medians as well as minima and maxima. For the CIs of the ROC curves and area under the ROC curves (AUROCs) we employed ‘pROC’s built-in boot.n function (also with B = 999 runs). Group-specific differences were evaluated by the Mann–Whitney-U-test (*P < 0.05; **P < 0.001). Kendall’s correlation plot (Fig. 1) was drawn with ‘rattle’ [see Murdoch and Chow (1996) for details]. We computed point-biserial correlation coefficients r pb by ‘R’s ‘ltm’ (Rizopoulos 2010) package, the significance levels thereof by the method proposed by Israel (Israel 2008). ROC curve and AUROC calculations were performed using ‘R’s ‘pROC’ package. Since we presumed that marker models including combinations of different amino acids and/or CEA might be superior over single amino acids and/or CEA regarding their selectivity, we also evaluated combined models. For this purpose, we selected amino acids significantly different between the groups (cf. Table 2) and, following power transformation (In-Kwon and Johnson 2000), performed a Principal Component Analysis [PCA, Eigenvalue <1, applying ‘R’s ‘princomp’ function of the ‘stats’ package (Venables and Ripley 2002)] thereon to minimize collinearities and to set up principal components for subsequent regression analysis and AUROC evaluation. We used combinations of these principal components as well as the concentrations of the significantly differing amino acids together with CEA in binary logistic regression modeling [package ‘BMA’, selection of the best fitting models and penalizing overfitting via the Bayesian Information Criterion (BIC), observing the degree of multicollinearity via the Variance Inflation Factor (VIF), model validation via the CAR score (defined as ‘the marginal correlations adjusted for correlation among explanatory variables’ with the acronym ‘CAR’ as an abbreviation for ‘Correlation-Adjusted (marginal) coRrelation’) applying package ‘care’ for the respective number of predictors, cf. Zuber and Strimmer (2010b)] to build predictors and to compare their selectivity with the selectivity of CEA alone by AUROC analysis in turn. Pvalues for the comparison of AUCs are computed as proposed by DeLong et al. (1988) for AUCs generated by the ‘pROC’ package [cf. Robin et al. (2010)]. ‘Best’ thresholds determined by the Youden index were computed using the ‘pROC’ package according to the method suggested by Perkins and Schisterman (2006). Non-inferiority and superiority testing thereafter was performed applying bootstrap techniques (B = 999, package ‘boot’) on ΔAUROC, constructing CIs thereof, and testing for embracement of Δ0 by the ΔAUROC’s CI according to the methods proposed by Liu et al. (2006), Tunes da Silva et al. (2009), and Lesaffre (2008) at a predefined δ L level of 5% which we considered to be medically reasonable (Mascha 2010) designing the study.
Fig. 1.
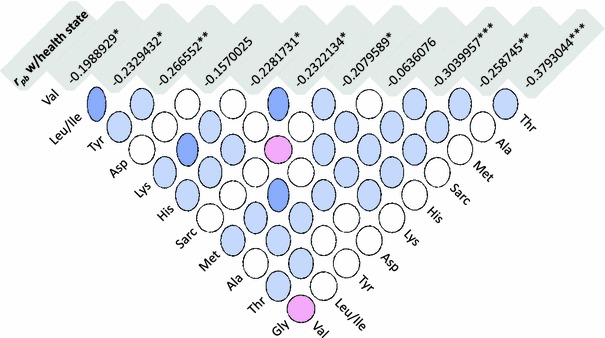
Correlation matrix plot of the amino acid concentrations [Kendall’s τb, circle denotes low correlation and oval denotes high correlation, positive, negative, and neutral correlations are displayed blue, red, and white, respectively (see Murdoch and Chow (1996) for details)] to display the mutual collinearities between amino acid concentrations. The top line (shaded) shows point biserial correlations r pb and significance levels for the correlation of the respective amino acid with the health state (*P < 0.05; **P < 0.01; ***P < 0.001)
Table 2.
Median, minimum, maximum, and the 95% CI of the median for amino acids in colon cancer patients and controls
| Colon cancer patients (n = 59) [μmol/l]a | Controls (n = 58) [μmol/l]a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Median | Range | Median CI 95% | Median | Range | Median CI 95% | |||||
| Minimum | Maximum | Low | High | Minimum | Maximum | Low | High | |||
|
Glutamine (Gln) (CID 5961) |
0.72 | 0.27 | 1.62 | 0.72 | 0.72 | 0.69 | 0.42 | 1.86 | 0.69 | 0.69 |
|
Lysine (Lys)* (CID 5962) |
232.41 | 136.62 | 481.02 | 231.6 | 233.25 | 278.52 | 133.56 | 133.56 | 277.56 | 279.48 |
|
Hydroxyproline (OH-Prol) (CID 5810) |
365.85 | 222.81 | 731.04 | 364.95 | 366.78 | 405.99 | 232.65 | 954.00 | 405.15 | 406.8 |
|
Pipecolic acid (PiPA) (CID 849) |
94.32 | 64.62 | 228.27 | 228.27 | 94.53 | 99.78 | 61.59 | 178.02 | 99.54 | 100.05 |
|
Abscisic acid (Aba) (CID 5280896) |
6.90 | 2.25 | 21.24 | 6.87 | 6.93 | 7.20 | 3.09 | 14.67 | 7.17 | 7.23 |
|
Alaninea (Ala)** (CID 5950) |
238.95 | 124.08 | 408.24 | 238.32 | 239.55 | 282.66 | 174.18 | 453.96 | 281.67 | 283.62 |
|
Argininea (Arg) (CID 6322) |
71.52 | 38.79 | 129.27 | 71.34 | 71.70 | 77.58 | 36.45 | 139.08 | 77.43 | 77.76 |
|
Aspartic acida (Asp)* (CID 5960) |
328.26 | 89.07 | 1821.69 | 324.3 | 332.22 | 538.74 | 75.75 | 1300.23 | 534.75 | 542.76 |
|
Carnosin (Carn) (CID 439224) |
1.29 | 0.03 | 3.72 | 1.29 | 1.29 | 1.17 | 0.30 | 3.06 | 1.14 | 1.17 |
|
Citrullinea (Cit) (CID 9750) |
22.77 | 10.80 | 51.75 | 22.68 | 22.83 | 25.44 | 12.54 | 40.47 | 25.38 | 25.50 |
|
Glutamic acida (Glu) (CID 33032) |
129.99 | 86.82 | 187.38 | 129.75 | 130.23 | 127.32 | 77.25 | 207.66 | 126.99 | 127.65 |
|
Glycinea (Gly)*** (CID 750) |
151.35 | 87.51 | 269.13 | 151.08 | 151.62 | 180.96 | 107.64 | 359.16 | 180.06 | 181.89 |
|
Histidine (His)*** (CID 6274) |
79.80 | 39.30 | 221.19 | 79.62 | 80.01 | 96.15 | 62.01 | 200.91 | 95.91 | 96.39 |
|
Leucinea/Isoleucine (Leu/Ile)** (CID 6106/CID 6306) |
114.18 | 68.67 | 232.47 | 113.82 | 114.54 | 139.74 | 75.96 | 272.31 | 139.41 | 140.1 |
|
Methyl-Histidine (MeHis) (CID 64969) |
9.03 | 3.66 | 49.77 | 8.97 | 9.09 | 8.94 | 3.57 | 22.92 | 8.91 | 9.00 |
|
Methioninea (Met)* (CID 6137) |
14.58 | 2.97 | 78.60 | 14.55 | 14.64 | 16.92 | 9.87 | 35.28 | 16.86 | 16.98 |
|
Ornithinea (Orn) (CID 6262) |
53.43 | 31.83 | 88.08 | 53.34 | 53.55 | 55.65 | 26.49 | 108.36 | 55.53 | 55.77 |
|
Phenylalaninea (Phe) (CID 994) |
58.65 | 37.14 | 111.57 | 58.50 | 58.83 | 61.23 | 37.38 | 132.78 | 61.08 | 61.38 |
|
Proline (Pro) (CID 8988) |
186.15 | 98.13 | 408.87 | 185.52 | 186.78 | 178.92 | 112.38 | 520.41 | 178.35 | 179.52 |
|
Sarcosine (Sarc)* (CID 1088) |
85.20 | 55.44 | 178.29 | 84.99 | 85.38 | 99.90 | 51.09 | 215.07 | 99.51 | 100.32 |
|
Serine (Ser) (CID 5951) |
96.90 | 50.67 | 163.29 | 96.69 | 97.14 | 92.79 | 54.63 | 205.92 | 92.55 | 93.03 |
|
Taurine (Tau) (CID 1123) |
2.46 | 1.23 | 5.19 | 2.46 | 2.49 | 2.31 | 0.90 | 5.43 | 2.28 | 2.31 |
|
Threonine (Thr)** (CID 6288) |
11.73 | 6.09 | 22.68 | 11.7 | 11.76 | 14.01 | 7.35 | 25.65 | 13.98 | 14.04 |
|
Tryptophan (Trp) (CID 6305) |
7.65 | 3.57 | 21.21 | 7.62 | 7.65 | 7.68 | 3.09 | 17.28 | 7.65 | 7.68 |
|
Tyrosinea (Tyr)* (CID 6057) |
46.08 | 27.27 | 69.96 | 45.96 | 46.23 | 50.52 | 32.64 | 92.79 | 50.40 | 50.64 |
|
Valinea (Val)* (CID 6287) |
231.99 | 132.99 | 364.50 | 231.51 | 232.50 | 260.49 | 158.49 | 401.97 | 260.07 | 260.94 |
Significance of group differences is given by * for P < 0.05, ** for P < 0.01 and *** for P < 0.001 (Wilcoxon-test); Italics show significant deviation from normal distribution (Anderson–Darling-test, computed on the original data). Median, minimum, maximum, and the 95% CI of the median are computed on the bootstrapped data (B = 999). Concentrations are μmol/l for amino acids quantified via internal standard (marked by a) and relative for all others (cf. Brauer et al. 2011). Metabolites are identified by their PubChem Compound ID (CID)
Results
For our investigations we collected serum samples of 59 (37m/22f) colorectal carcinoma patients and 58 [26m/32f; P = not significant (n.s.)] healthy controls. Age, UICC (Union Internationale Contre le Cancer) stagings of the patients, and CEA concentrations are displayed in Table 1.
Table 1.
Baseline characteristics
| Baseline characteristics | Colon cancer patients | Healthy controls |
|---|---|---|
| Male/Female | 37/22 | 26/32 |
| Age (years; median, 2.5th–97.5th) | 59 (45–90) | 58 (38–75) |
| CEA (ng/ml; Median, 2.5th–97.5th) | 3.5 (0.9–2387.0) | 1.7 (0.6–3.7) |
| UICC staging (stage 1/2/3/4) | 5/18/20/16 | – |
Baseline characteristics of the colorectal cancer and the healthy control group. Both groups differed highly significant in their CEA serum concentrations (P < 0.001)
Descriptives
To generate robust estimators for the amino acid concentrations in the colorectal carcinoma patients and healthy controls covering the value range and the 95% confidence interval of the median for comparative studies, we applied bootstrapping techniques—the resampling results are summarized in Table 2 and displayed separately for both groups. In total, we found 19 of 26 amino acids decreased (11 thereof significantly) and 7 amino acids increased (none significantly) in colorectal carcinoma patients compared to healthy controls. Several amino acids were non-normally distributed across both study groups (Table 2).
Correlations
To evaluate the correlation of the significantly different amino acids with the health state and with each other, we computed point-biserial correlations (r
pb) and Kendall’s τb, respectively. r
pb ranged between –0.38 and 0.10 (with P < 0.05 for
Modelling
As we conjectured that the use of combinatory markers including multiple amino acids and/or CEA could yield additive effects and outclass single amino acids and/or CEA (Schneider et al. 2002; Wild et al. 2010), we built combined models to evaluate their selectivity in comparison with single amino acids and CEA. To control for multicollinearity, which is a significant restraint for logistic regression (Leigh 1988), we performed PCA following power transformation on the significantly different amino acids and processed the resulting principal components (PCs) as well as the corresponding single amino acid concentrations and CEA by binary logistic regression modeling. The best PCA-based model comprised CEA and PC1, the best amino acid concentration-based model CEA, Glycine, and Tyrosine with a slightly better fit (ΔBIC–2.8).
Receiver–operator-characteristics analysis
We used the single amino acid concentrations as well as the predictors gained from the two best-fitting models for AUROC analysis. The results are displayed in Table 3 for the amino acids and in Fig. 2 for the predictors compared to CEA. The best-discriminating model comprised CEA, Glycine, and Tyrosine (AUROC 0.878, 95% CI 0.815–0.941), followed by the model comprising CEA and PC1 (AUROC 0.844, 95% CI 0.773–0.916), CEA alone (AUROC 0.794, 95% CI 0.712–0.877) and Glycine (AUROC 0.707, 95% CI 0.613–0.801) as the best discriminating single amino acid. Both predictor models were not significantly different, the CEA, Glycine, and Tyrosine model, however, differed highly significant from CEA alone (P = 0.015).
Table 3.
Amino acid AUROCs and their 95% CIs for the differentiation of colon cancer patients and controls
| Amino acids | AUC | 95% CI | |
|---|---|---|---|
| Low | High | ||
| Glycine | 0.707 | 0.613 | 0.801 |
| Histidine | 0.691 | 0.593 | 0.788 |
| Alanine | 0.677 | 0.580 | 0.774 |
| Leucine/Isoleucine | 0.653 | 0.551 | 0.754 |
| Threonine | 0.645 | 0.544 | 0.746 |
| Tyrosine | 0.636 | 0.536 | 0.737 |
| Methionine | 0.633 | 0.532 | 0.735 |
| Valine | 0.625 | 0.521 | 0.728 |
| Lysine | 0.624 | 0.522 | 0.725 |
| Aspartate | 0.622 | 0.520 | 0.724 |
| Sarcosine | 0.617 | 0.515 | 0.719 |
| Arginine | 0.594 | 0.490 | 0.697 |
| Hydroxyproline | 0.574 | 0.467 | 0.681 |
| Pipecholic | 0.568 | 0.463 | 0.673 |
| Phenylalanine | 0.568 | 0.463 | 0.672 |
| Carnosin | 0.558 | 0.452 | 0.663 |
| Citrullin | 0.554 | 0.447 | 0.660 |
| Methylhistidine | 0.536 | 0.430 | 0.642 |
| Glutamine | 0.530 | 0.424 | 0.635 |
| Ornithine | 0.527 | 0.420 | 0.634 |
| Taurine | 0.523 | 0.414 | 0.633 |
| Abscisic acid | 0.521 | 0.415 | 0.627 |
| Tryptophane | 0.506 | 0.400 | 0.612 |
| Serine | 0.506 | 0.399 | 0.612 |
| Glutamine | 0.485 | 0.378 | 0.591 |
| Proline | 0.463 | 0.357 | 0.569 |
AUROCs and 95% CIs for the different amino acids with respect to the discriminatory power between colon cancer patients and healthy controls
Fig. 2.
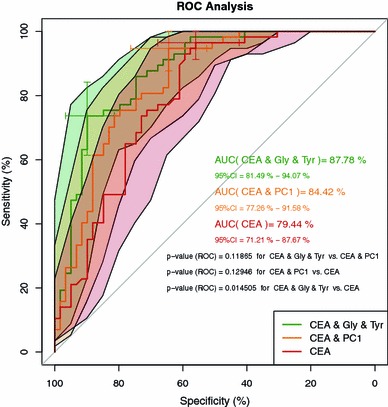
ROC curves, 95% confidence bands (B = 999) and AUROCs for CEA (red), the model consisting of CEA and principal component 1 (PC1 orange), and the model consisting of CEA, glycine, and tyrosine (green). Sensitivity and specificity and their 95% confidence intervals (B = 999) are given for the best thresholds and denoted by (☩). P-values between AUCs are computed according to DeLong et al. (1988) as described in the Sect. 2
Non-inferiority and superiority testing
Since significant difference is not an adequate measure of non-inferiority or superiority, we performed sequential testing for both with an a priori-defined acceptance criterion (equivalence limit) of δ L = 5% ΔAUROC. We computed the lower and upper limits of the (100 − 2 δ L)% bootstrap confidence interval (0.0239–0.1422) of the estimated ΔAUROC (B = 999) as proposed by Liu et al. (2006). Referring to Mascha (2010) we deduced non-inferiority in a first step, and—as the lower CI of ΔAUROC is >0—inferred superiority of the model containing CEA, Glycine, and Tyrosine over CEA alone thereafter.
Discussion
We found the concentrations of 11 out of 26 serum amino acids as significantly different between CRC patients and healthy controls. To additionally evaluate the diagnostic potential of our results, we applied a bioinformatic pipeline comprising common standard test statistics as well as AUROC analysis followed by non-inferiority/superiority testing and found a model including CEA, glycine, and tyrosine as best discriminating and superior to CEA alone with an AUROC of 0.878 (95% CI 0.815–0.941).
The rationale of our study was to detect and evaluate alterations of the amino acid profile in CRC patients as a potential source of complementary metabolic markers by means of standardized preanalytics, a routinely applicable analytical method, and a computational procedure, which allows statistically sound statements (Walker and Nowacki 2010) on the diagnostic surplus value.
The recent methodological advancements in the rapidly emerging field of clinical metabolomics preceded numerous studies investigating metabolic signatures of colorectal cancer. A variety of different mass-spectrometric platforms has been applied and yielded different marker analytes. Comparable to our results, Qiu et al. (2009) also found serum lysine, leucine, threonine decreased, valine, and tyrosine decreased [the latter with both applied techniques, Gas chromatography time-of-flight mass spectrometry (GC-TOF MS) and Ultraperformance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOF MS)], Ma et al. (2009) valine, threonine and glycine significantly decreased in CRC patients using GC-MS. Denkert et al. (2008) found in a comprehensive study alanine, methionine, threonine, leucine, isoleucine, valine, and less significantly also glycine and lysine (GC-TOF MS) increased in human CRC tissues, Chan et al. (2009) as well as Tessem et al. (2010) [both with High-resolution magic angle spinning nuclear magnetic resonance spectrometry (HR-MAS NMR)] and Hirayama et al. (2009) [Capillary electrophoresis time-of-flight mass spectrometry (CE-TOF MS)] found glycine, and Ong et al. (2010) [Gas chromatography mass spectrometry (GC-MS)] methionine and tyrosine elevated in CRC tissue samples, whereas Ma et al. (2010) (also with GC-MS) comparing pre-/postoperative samples found serum valine decreased and tyrosine increased after surgery. Another study by Qiu et al. (2010) focused on urinary metabolites and revealed lower histidine concentrations in CRC urine samples. The comparability of these studies among each other and with our study is limited due to different analytical techniques, sample material as well as provenience, and patient classificators. Actually, there is no published study comparing MS/MS-based serum amino acid profiles of CRC patients and controls (Wang et al. 2010). Additionally, the investigations of Ma et al. (2010) and Qiu (2009) primarily aimed at the detection of significant differences of the focused metabolites and not at the evaluation of their diagnostic selectivity by canonical methods like (Area under the) receiver–operator-characteristics [(AU)ROC] analysis, as e.g. Ritchie et al. (2010) performing Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS) experiments and Chan et al. (2009) performing HR-MAS NMR analyses did for ultra long-chain fatty acids and tissue samples, respectively. In both of these last-mentioned studies the discriminating analytes were, as “standalone”-markers or as OPLS-DA-models, evaluated independently from the conventional tumor marker CEA and their equivalence was not surveyed.
Despite the limited comparability of the above-mentioned investigations with our study, the eleven significantly decreased amino acids we detected were—except for sarcosine, and with varying significance—also identified as marker metabolites for CRC in other studies.
The best discriminating amino acid glycine (AUROC 0.707), which was also found by Ma et al. (2009) and Tessem et al. (2010), is an important intermediate in the folate metabolism, which is especially altered in colon cancer (Stover and MacFarlane 2008). Due to its recently demonstrated bowel-protective effects and its easy applicability it is also a highly interesting candidate for therapeutic approaches (de Aguiar Picanco et al. 2011).
In biomarker research, it is a common practice to analytically determine marker molecules, compute the statistical significance of their differences between groups and to perform e.g. PCA-based analyses resulting in rotated, non-linear, and non-retraceable (and therefore non-comparable), but stunningly separating “component plots”, leaving the reader alone with their interpretation. Only few studies implemented AUROC analyses (Chan et al. 2009; Ritchie et al. 2010) and none evaluated non-inferiority/superiority. It is a core task and competence of laboratory medicine, not only to investigate potential biomarkers, but also to generate lucid test characteristics, which allow the comprehensible interpretation of their diagnostic value and their translation into clinical practice. Therefore, we augmented our study by four add-ons: first, we generated robust estimates for summary statistics of the amino acid concentrations to facilitate comparability with other studies despite the relatively small number of samples. Second, we integrated the anyway available CEA concentrations into BIC-based logistic regression modeling to utilize its auxiliary selectivity. Third, we compared amino acid concentrations themselves with the typically used principal components thereof and—fourth—specified the non-inferiority/superiority of our models compared to the conventional tumor marker CEA based on AUROC analyses.
Tyrosine was identified as marker metabolite by Qiu et al. (2009), Ma et al. (2010, 2009), Vecer et al. (1998), and Ong et al. (2010) and associated with cancer-related alterations of the TCA cycle by Hirayama et al. (2009), but in contrast to glycine, it displayed inferior selectivity with an AUROC of 0.636 in our study. However, combined with CEA and glycine, it surprisingly is part of the overall best discriminating model, suggesting that its contribution to the selectivity of the model features aspects, which are different from or stronger than other amino acids, even if the latter (e.g. histidine or alanine) individually are stronger discriminators. Interestingly, the model comprising the two amino acid concentrations was slightly more selective than that based on PCA, inferring that variance-based techniques—despite their frequent application—might not always yield optimal classifications.
Even though the AUROCs of the best-discriminating amino acids almost reach the range of CEA, they are of limited use as single discriminators. In combination with the conventional tumor marker CEA, which had an AUROC (0.794) comparable to the recent literature (Wild et al. 2010), glycine and tyrosine, however, introduced a surplus of 8%, suggesting that—regarding their AUROCs of 0.707 and 0.636 as single markers—their principal value might be of additive nature and be missed when routinely available CEA concentrations are not taken into account.
Besides these emboldening results, there are some limitations to consider: whereas the determination of glycine and tyrosine is based on isotope labeled internal standards and of quantitative nature, the concentrations of several other amino acids (cf. Table 2) have to be considered as relative. For AUROC analyses, this might not be relevant, since deviations affect both, controls and patients. Nonetheless, direct comparisons with other studies are limited for these parameters. Another criticism might arise from the considerable overlap of the amino acid concentrations between patients and controls as outlined by Issaq et al. (2011) reviewing a metabolomic tumor marker study of Kim et al. (2010) and denying potential use of the analytes displayed there as biomarkers or for population studies. However, applying the same standards on established tumor markers, even CEA would inevitably fail. To minimize this inherent overlap problem, we generated multiparametric (and thereby multidimensional) models (Robin et al. 2009; Wild et al. 2010). A third constraint is implicated in our dichotomous study design consisting of two well-separated cohorts, colorectal tumor patients on the one and healthy controls on the other side. This pilot study design is suited to maximize differences between both groups to enhance pre-test probability, but it cannot deliver differentiation between CRC and e.g. inflammatory bowel diseases. Along with the size of the cohort differences the statistical power is diminishing and the number of study participants must be raised. With our preliminary study we tried to find an optimal significance of the results balancing study size and study group dissimilarity.
Concluding remarks
We analyzed 117 highly standardized serum samples and generated amino acid profiles by applying a relatively simple high-throughput mass-spectrometric technique. In addition to the common reporting of the significance of the concentration difference between groups and PCA-based modeling, our bioinformatic pipeline included (AU)ROC analyses, integrative BIC-based logistic regression modeling and non-inferiority/superiority testing to exemplarily and numerically determine the diagnostic surplus value for the clinician and to avoid the ambiguity remaining with a significant difference. In comparison with the conventional tumor marker for colorectal cancer CEA, our model additionally containing glycine and tyrosine was superior. Further large-scale studies are necessary to elucidate the potential of this model also to discriminate between cancer and potential differential diagnoses.
Acknowledgments
The authors thank Lars Schinkel for capably performing the LC–MS/MS measurements at the Institute of Laboratory Medicine of the University Hospital Leipzig, Germany. The authors also thank Xavier Robin for his skillful support adapting the pROC package to their needs.
Conflict of interest
All authors state that no conflict of interest exists.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- (i)FOBT
(immunological) Fecal occult blood test
- (AU)ROC
(Area under the) receiver–operator-characteristics
- AUC
Area under the curve
- CRC
Colorectal cancer
- TCA
Tricarboxylic acid
- MS/MS
Tandem mass spectrometry
- GC-MS
Gas chromatography mass spectrometry
- HR-MAS NMR
High-resolution magic angle spinning nuclear magnetic resonance spectrometry
- FTICR-MS
Fourier transform ion cyclotron resonance mass spectrometry
- CE-TOF MS
Capillary electrophoresis time-of-flight mass spectrometry
- GC-TOF MS
Gas chromatography time-of-flight mass spectrometry
- UPLC-QTOF MS
Ultraperformance liquid chromatography-quadrupole time-of-flight mass spectrometry
- PCA
Principal component analysis
- (O)PLS-DA
(Orthogonal) partial least squares discriminant analysis.
Footnotes
Alexander Benedikt Leichtle and Jean-Marc Nuoffer contributed equally to this paper.
References
- Baumann S, Ceglarek U, Fiedler GM, Lembcke J, Leichtle A, Thiery J. Standardized approach to proteome profiling of human serum based on magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clinical Chemistry. 2005;51:973–980. doi: 10.1373/clinchem.2004.047308. [DOI] [PubMed] [Google Scholar]
- Brauer R, Leichtle A, Fiedler G, Thiery J, Ceglarek U. Preanalytical standardization of amino acid and acylcarnitine metabolite profiling in human blood using tandem mass spectrometry. Metabolomics. 2011;7(3):344–352. doi: 10.1007/s11306-010-0256-1. [DOI] [Google Scholar]
- Bretthauer M. Evidence for colorectal cancer screening. Best Practice and Research Clinical Gastroenterology. 2010;24:417–425. doi: 10.1016/j.bpg.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Cardoso J, Boer J, Morreau H, Fodde R. Expression and genomic profiling of colorectal cancer. Biochimica et Biophysica Acta. 2007;1775:103–137. doi: 10.1016/j.bbcan.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Carpenter J, Bithell J. Bootstrap confidence intervals: When, which, what? A practical guide for medical statisticians. Statistics in Medicine. 2000;19:1141–1164. doi: 10.1002/(SICI)1097-0258(20000515)19:9<1141::AID-SIM479>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Cascante M, Marin S. Metabolomics and fluxomics approaches. Essays Biochem. 2008;45:67–81. doi: 10.1042/BSE0450067. [DOI] [PubMed] [Google Scholar]
- Ceglarek U, Leichtle A, Brugel M, Kortz L, Brauer R, Bresler K, Thiery J, Fiedler GM. Challenges and developments in tandem mass spectrometry based clinical metabolomics. Molecular and Cellular Endocrinology. 2009;301:266–271. doi: 10.1016/j.mce.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Ceglarek U, Muller P, Stach B, Buhrdel P, Thiery J, Kiess W. Validation of the phenylalanine/tyrosine ratio determined by tandem mass spectrometry: Sensitive newborn screening for phenylketonuria. Clinical Chemistry and Laboratory Medicine. 2002;40:693–697. doi: 10.1515/CCLM.2002.119. [DOI] [PubMed] [Google Scholar]
- Chan EC, Koh PK, Mal M, Cheah PY, Eu KW, Backshall A, Cavill R, Nicholson JK, Keun HC. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS) Journal of Proteome Research. 2009;8:352–361. doi: 10.1021/pr8006232. [DOI] [PubMed] [Google Scholar]
- Davison AC, Hinkley DV, editors. Bootstrap methods and their applications. Cambridge: Cambridge University Press; 1997. [Google Scholar]
- de Aguiar Picanco E, Lopes-Paulo F, Marques RG, Diestel CF, Caetano CE, de Souza MV, Moscoso GM, Pazos HM. l-arginine and glycine supplementation in the repair of the irradiated colonic wall of rats. International Journal of Colorectal Disease. 2011;26:561–568. doi: 10.1007/s00384-011-1154-3. [DOI] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- Denkert C, Budczies J, Weichert W, Wohlgemuth G, Scholz M, Kind T, Niesporek S, Noske A, Buckendahl A, Dietel M, Fiehn O. Metabolite profiling of human colon carcinoma–deregulation of TCA cycle and amino acid turnover. Molecular Cancer. 2008;7:72. doi: 10.1186/1476-4598-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DESTATIS, Statistisches Bundesamt Deutschland. (2009). Sterbefälle insgesamt 2009 nach den 10 häufigsten Todesursachen der International Statistical Classification of Diseases and Related Health Problems (ICD-10). http://www.destatis.de/.
- Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrometry Reviews. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, van Rossum LG, van Turenhout ST, Malminiemi O, Sturgeon C, Lamerz R, Nicolini A, Haglund C, Holubec L, Fraser CG, Halloran SP. Use of faecal markers in screening for colorectal neoplasia: A European group on tumor markers position paper. International Journal of Cancer. 2011;128:3–11. doi: 10.1002/ijc.25654. [DOI] [PubMed] [Google Scholar]
- Fiedler GM, Ceglarek U, Lembcke J, Baumann S, Leichtle A, Thiery J. Application of mass spectrometry in clinical chemistry and laboratory medicine. Journal of Laboratory Medicine. 2004;28:185–194. [Google Scholar]
- Fiedler GM, Leichtle AB, Kase J, Baumann S, Ceglarek U, Felix K, Conrad T, Witzigmann H, Weimann A, Schutte C, Hauss J, Buchler M, Thiery J. Serum peptidome profiling revealed platelet factor 4 as a potential discriminating peptide associated with pancreatic cancer. Clinical Cancer Research. 2009;15:3812–3819. doi: 10.1158/1078-0432.CCR-08-2701. [DOI] [PubMed] [Google Scholar]
- Fiehn O, Robertson D, Griffin J, van der Werf M, Nikolau B, Morrison N, Sumner L, Goodacre R, Hardy N, Taylor C, Fostel J, Kristal B, Kaddurah-Daouk R, Mendes P, van Ommen B, Lindon J, Sansone S-A. The metabolomics standards initiative (MSI) Metabolomics. 2007;3:175–178. doi: 10.1007/s11306-007-0070-6. [DOI] [Google Scholar]
- Fox J, Weisberg S, editors. An R Companion to applied regression. New Jersey: Sage Publications Inc.; 2011. [Google Scholar]
- Graham JW. Rattle: A data mining GUI for R. The R Journal. 2009;1:44–55. [Google Scholar]
- Gross, J. (2006). nortest: Tests for normality. http://cran.r-project.org/web/packages/nortest/.
- Harder U, Koletzko B, Peissner W. Quantification of 22 plasma amino acids combining derivatization and ion-pair LC-MS/MS. Journal of Chromatography B. 2011;879:495–504. doi: 10.1016/j.jchromb.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Herszenyi L, Tulassay Z. Epidemiology of gastrointestinal and liver tumors. European Review for Medical and Pharmacological Sciences. 2011;14:249–258. [PubMed] [Google Scholar]
- Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, Esumi H, Soga T. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Research. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- In-Kwon Y, Johnson RA. A new family of power transformations to improve normality or symmetry. Biometrika. 2000;87:954–959. doi: 10.1093/biomet/87.4.954. [DOI] [Google Scholar]
- Israel D. Data analysis in business research: A step-by-step nonparametric approach. London: SAGE Publications Ltd.; 2008. [Google Scholar]
- Issaq HJ, Waybright TJ, Veenstra TD. Cancer biomarker discovery: Opportunities and pitfalls in analytical methods. Electrophoresis. 2011;32:967–975. doi: 10.1002/elps.201000588. [DOI] [PubMed] [Google Scholar]
- Kim J-W, Lee G, Moon S-M, Park M-J, Hong S, Ahn Y-H, Kim K-R, Paik M-J. Metabolomic screening and star pattern recognition by urinary amino acid profile analysis from bladder cancer patients. Metabolomics. 2010;6:202–206. doi: 10.1007/s11306-010-0199-6. [DOI] [Google Scholar]
- Leigh JP. Assessing the importance of an independent variable in multiple regression: Is stepwise unwise? Journal of Clinical Epidemiology. 1988;41:669–677. doi: 10.1016/0895-4356(88)90119-9. [DOI] [PubMed] [Google Scholar]
- Lesaffre E. Superiority, equivalence, and non-inferiority trials. Bulletin of the NYU Hospital for Joint Diseases. 2008;66:150–154. [PubMed] [Google Scholar]
- Liu J-P, Ma M-C, Wu C-y, Tai J-Y. Tests of equivalence and non-inferiority for diagnostic accuracy based on the paired areas under ROC curves. Statistics in Medicine. 2006;25:1219–1238. doi: 10.1002/sim.2358. [DOI] [PubMed] [Google Scholar]
- Ma Y, Liu W, Peng J, Huang L, Zhang P, Zhao X, Cheng Y, Qin H. A pilot study of gas chromatograph/mass spectrometry-based serum metabolic profiling of colorectal cancer after operation. Molecular Biology Reports. 2010;37:1403–1411. doi: 10.1007/s11033-009-9524-4. [DOI] [PubMed] [Google Scholar]
- Ma YL, Liu WJ, Peng JY, Zhang P, Chen HQ, Qin HL. Study on specific metabonomic profiling of serum from colorectal cancer patients by gas chromatography-mass spectrometry. Zhonghua Wei Chang Wai Ke Za Zhi. 2009;12:386–390. [PubMed] [Google Scholar]
- Mamas M, Dunn WB, Neyses L, Goodacre R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Archives of Toxicology. 2011;85:5–17. doi: 10.1007/s00204-010-0609-6. [DOI] [PubMed] [Google Scholar]
- Mascha EJ. Equivalence and noninferiority testing in anesthesiology research. Anesthesiology. 2010;113:779–781. doi: 10.1097/ALN.0b013e3181ec6212. [DOI] [PubMed] [Google Scholar]
- Mueller P, Schulze A, Schindler I, Ethofer T, Buehrdel P, Ceglarek U. Validation of an ESI-MS/MS screening method for acylcarnitine profiling in urine specimens of neonates, children, adolescents and adults. Clinica Chimica Acta. 2003;327:47–57. doi: 10.1016/S0009-8981(02)00327-3. [DOI] [PubMed] [Google Scholar]
- Murdoch DJ, Chow ED. A graphical display of large correlation matrices. The American Statistician. 1996;50:178–180. [Google Scholar]
- Ong, E. S., Zou, L., Li, S., Cheah, P. Y., Eu, K. W., & Ong, C. N. (2010). Metabolic profiling in colorectal cancer reveals signature metabolic shifts during tumorigenesis. Molecular and Cellular Proteomics. doi:10.1074/mcp.M900551-MCP200. [DOI] [PubMed]
- Perkins NJ, Schisterman EF. The inconsistency of “optimal” cutpoints obtained using two criteria based on the receiver operating characteristic curve. American Journal of Epidemiology. 2006;163:670–675. doi: 10.1093/aje/kwj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Cai G, Su M, Chen T, Liu Y, Xu Y, Ni Y, Zhao A, Cai S, Xu LX, Jia W. Urinary metabonomic study on colorectal cancer. Journal of Proteome Research. 2010;9:1627–1634. doi: 10.1021/pr901081y. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, Ni Y, Zhao A, Xu LX, Cai S, Jia W. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. Journal of Proteome Research. 2009;8:4844–4850. doi: 10.1021/pr9004162. [DOI] [PubMed] [Google Scholar]
- Raftery, A., Hoeting, J., Volinsky, C., Painter, I. & Yeung, K. Y. (2010). Bayesian Model Averaging, CRAN. Package for Bayesian model averaging for linear models, generalizable linear models and survival models (cox regression). http://cran.r-project.org/web/packages/BMA/.
- Ritchie SA, Ahiahonu PW, Jayasinghe D, Heath D, Liu J, Lu Y, Jin W, Kavianpour A, Yamazaki Y, Khan AM, Hossain M, Su-Myat KK, Wood PL, Krenitsky K, Takemasa I, Miyake M, Sekimoto M, Monden M, Matsubara H, Nomura F, Goodenowe DB. Reduced levels of hydroxylated, polyunsaturated ultra long-chain fatty acids in the serum of colorectal cancer patients: Implications for early screening and detection. BMC Medicine. 2010;8:13. doi: 10.1186/1741-7015-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizopoulos, D. (2010). ltm: Latent trait models under IRT, CRAN. http://cran.r-project.org/web/packages/ltm/.
- Robin X, Turck N, Hainard A, Lisacek F, Sanchez JC, Muller M. Bioinformatics for protein biomarker panel classification: What is needed to bring biomarker panels into in vitro diagnostics? Expert Review of Proteomics. 2009;6:675–689. doi: 10.1586/epr.09.83. [DOI] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin, X., Turck, N., Hainard, A., Tiberti, N., Lisacek, F., Sanchez, J. & Müller, M. (2010). pROC—display and analyze ROC curves: Tools for visualizing, smoothing and comparing receiver operating characteristic (ROC curves). http://cran.r-project.org/web/packages/pROC/.
- Schneider J, Bitterlich N, Velcovsky HG, Morr H, Katz N, Eigenbrodt E. Fuzzy logic-based tumor-marker profiles improved sensitivity in the diagnosis of lung cancer. International Journal of Clinical Oncology. 2002;7:145–151. doi: 10.1007/s101470200021. [DOI] [PubMed] [Google Scholar]
- Shimwell NJ, Wei W, Wilson S, Wakelam MJ, Ismail T, Iqbal T, Johnson PJ, Martin A, Ward DG. Assessment of novel combinations of biomarkers for the detection of colorectal cancer. Cancer Biomark. 2010;7:123–132. doi: 10.3233/CBM-2010-0155. [DOI] [PubMed] [Google Scholar]
- Stephens M. Anderson–Darling test of goodness of fit, encyclopedia of statistical sciences. New York: John Wiley; 2006. [Google Scholar]
- Stover PJ, MacFarlane AJ. Mouse models to elucidate mechanisms of folate-related cancer pathologies. Nutrition Reviews. 2008;66(Supp 1):S54–S58. doi: 10.1111/j.1753-4887.2008.00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tanaka M, Ishigamori R. Biomarkers for colorectal cancer. International Journal of Molecular Sciences. 2010;11:3209–3225. doi: 10.3390/ijms11093209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R Development Core Team (2008). R: A language and environment for statistical computing, R foundation for statistical computing. http://www.r-project.org/.
- Tessem MB, Selnaes KM, Sjursen W, Trano G, Giskeodegard GF, Bathen TF, Gribbestad IS, Hofsli E. Discrimination of patients with microsatellite instability colon cancer using 1H HR MAS MR spectroscopy and chemometric analysis. Journal of Proteome Research. 2010;9:3664–3670. doi: 10.1021/pr100176g. [DOI] [PubMed] [Google Scholar]
- Tunes da Silva G, Logan BR, Klein JP. Methods for equivalence and noninferiority testing. Biology of Blood and Marrow Transplantation. 2009;15:120–127. doi: 10.1016/j.bbmt.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecer J, Kvapil M, Sprongl L, Kubatova H, Hoch J, Jech Z, Charvat J. Tissue amino acids in patients with colorectal carcinoma. Vnitrni Lekarstvi. 1998;44:192–194. [PubMed] [Google Scholar]
- Venables, W. N. & Ripley, B. D. (2002). Modern applied statistics with S, 4th edn. New York: Springer.
- Walker E, Nowacki AS. Understanding equivalence and noninferiority testing. Journal of General Internal Medicine. 2010;26:192–196. doi: 10.1007/s11606-010-1513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tso VK, Slupsky CM, Fedorak RN. Metabolomics and detection of colorectal cancer in humans: a systematic review. Future Oncology. 2010;6:1395–1406. doi: 10.2217/fon.10.107. [DOI] [PubMed] [Google Scholar]
- Wang P, Whiteaker JR, Paulovich AG. The evolving role of mass spectrometry in cancer biomarker discovery. Cancer Biology and Therapy. 2009;8:1083–1094. doi: 10.4161/cbt.8.12.8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild N, Andres H, Rollinger W, Krause F, Dilba P, Tacke M, Karl J. A combination of serum markers for the early detection of colorectal cancer. Clinical Cancer Research. 2010;16:6111–6121. doi: 10.1158/1078-0432.CCR-10-0119. [DOI] [PubMed] [Google Scholar]
- Zuber, V. & Strimmer, K. (2010a). Package “CAR estimation, variable selection, and regression” for R. http://cran.r-project.org/web/packages/care/.
- Zuber, V. & Strimmer, K. (2010b). Variable importance and model selection by decorrelation. http://arxiv.org/abs/1007.5516.


