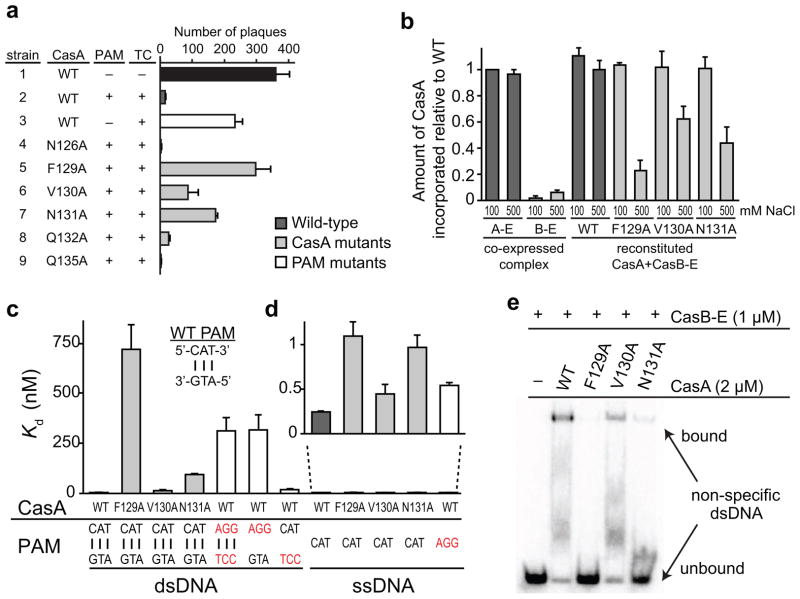
Figure 2.
CasA L1 is important for Cascade assembly and target binding. (a) Phage sensitivity of E. coli strains expressing a CRISPR/Cas system with either WT CasA or the indicated CasA mutant. Cas genes were co-expressed with a CRISPR containing spacers that are not complementary to lambda phage, a CRISPR containing spacers complementary to the coding strand of lambda phage genes J,O, R, and E but no PAM; or a CRISPR containing spacers complementary to the coding strand of lambda phage genes J,O, R, and E, and including a PAM. The presence (+) or absence (−) of a PAM and/or a targeting CRISPR (TC) is indicated. See also Table S1. The number of plaques were counted and values from three replicates were averaged. Error bars represent one standard deviation. (b) Incorporation defects for CasA L1 mutants. The amount of CasA relative to CasC was determined from denaturing protein gel band intensities and normalized against relative amount of CasA in low salt Cascade immunoprecipitation. Aggregate data from three replicates are shown, with error bars representing one standard deviation. For WT, F129A, V130A and N131A CasA, reconstituted CasA + CasBCDE-crRNA (CasB-E) complex was formed and washed in the presence of low (100 mM) or high (500 mM) concentrations of NaCl. (c–e) Target binding defects for CasA L1 and PAM mutants (PAM mutations labeled in red). (c–d) Bar graph plotting Kd values for (c) dsDNA or (c–d) ssDNA targets. Average values from three replicates are shown, with error bars representing one standard deviation. Targets according to nomenclature in Figure S1a are as follows: Bars 1–4: O1—O2 duplex; Bar 5: O3—O4 duplex; Bar 6: O3—O2 duplex; Bar 7: O1—O4 duplex; Bars 8–11: O1; Bar 12: O3. (e) Gel shift assay for non-specific O5—O6 dsDNA (Fig. S1a) target binding by WT and mutant CasA + CasBCDE-crRNA (CasB-E).