Figure 1.
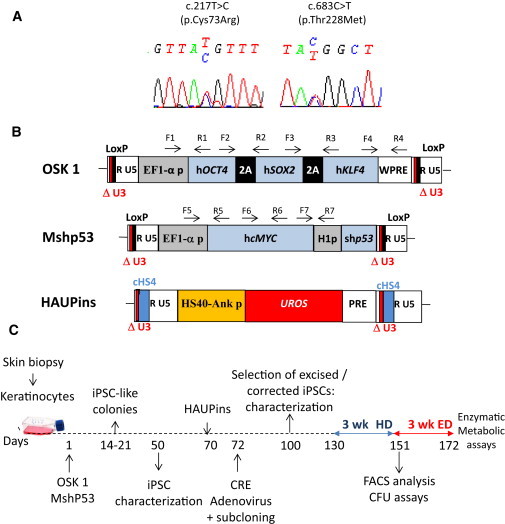
CEP Characterization and Schematic Drawing of iPSC Generation
(A) Genotyping of keratinocytes from a CEP-affected individual compound heterozygous for UROS mutations c.217T>C (p.Cys73Arg) and c.683C>T (p.Thr228Met).
(B) Schematic representation of the proviral form of the lentivectors used. OSK 1 is an excisable single polycistronic vector coexpressing OCT4, SOX2, and KLF4 cDNAs linked with the porcine teschovirus-1 2A sequence. Mshp53 coexpresses MYC and a shRNA against TP53; both vectors are flanked by loxP sites. HAUPins contains the UROS cDNA under the control of the erythroid chimeric HS-40 enhancer/ankyrin promoter. Arrows show the position of forward (F) and reverse (R) primers used for proviral integration analyses. The following abbreviations are used: HS40, enhancer of HBA1 (MIM 141800); Ank p, ankyrin promoter of ANKYRIN-1 (MIM 612641); UROS, cDNA of uroporphyrinogene III synthase; H1p, mammalian polymerase III H1 promoter; EF1α-p, elongation factor-1 alpha promoter; WPRE, woodchuck posttranscriptional regulatory element; PRE, a mutated WPRE sequence (WPREmut6); and cHS4, element of the chicken β-globin hypersensitivity site 4.
(C) Schematic strategy used for reprogramming human keratinocytes from a CEP-affected person and for HSC differentiation. The following abbreviations are used: HD, hematopoietic differentiation; and ED, erythroid differentiation.
