Figure 2.
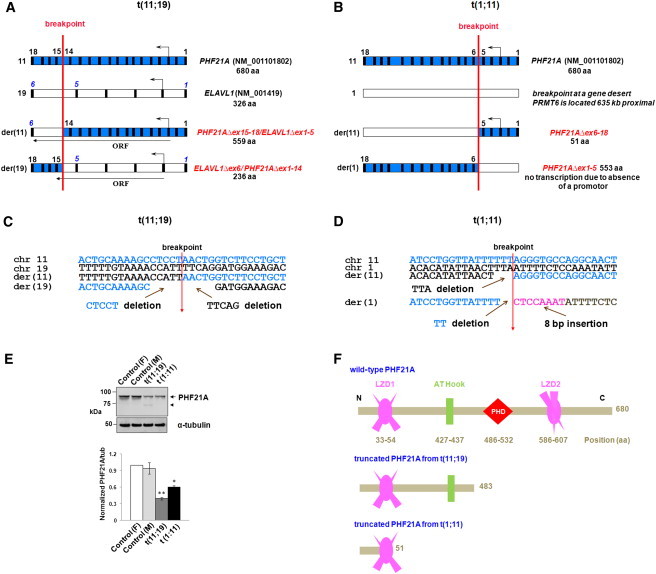
Mapping of the Breakpoints in Two Balanced Translocations
(A and B) Disruption of PHF21A by two translocations. A schematic diagram of the DGAP012 breakpoints (not to scale) shows, in blue, PHF21A on chromosome 11 (exons 1–18) and, in white, either (A) ELAVL1 on chromosome 19 (exons 1–6) or (B) chromosome 1. All exons are depicted as vertical black boxes, and the start and direction of transcription are indicated by an arrow above the corresponding exons. The translocation occurred at the site of the red vertical line. In DGAP012, the translocation produced der(11) and der(19) chromosomes that encode potential fusion transcripts (A). Note that the stop codon of the potential PHF21AΔex15-18/ELAVL1Δex1-5 fusion gene is at the same location as wild-type ELAVL1 because there is no frameshift, whereas ELAVL1Δex6/ PHF21AΔex1-14 has a frameshift with a premature stop codon in the new exon 6 (equivalent to exon 15 of PHF21A). The translocation in MCN1762 does not predict any potential fusion product because the breakpoint on chromosome 1 is located in a gene desert.
(C and D) Genomic DNA sequence from the normal chromosomes and at the breakpoints on derivative chromosomes. In DGAP012, at the der(19) breakpoint, there was a 5 nt CTCCT deletion of chromosome 11 sequence and a 5 nt TTCAG deletion of chromosome 19 sequence, whereas at the der(11) breakpoint, there was no loss or gain of sequence (C). In MCN1762, the junction sequences revealed a 3 bp TTA deletion of chromosome 1 sequence on der(11) and a 2 bp TT deletion of chromosome 11 sequence and an 8 bp CTCCAAAT insertion on der(1). Details of the mapping of both breakpoints in DGAP012 and MCN1762 are described in Figure S1 as well as in the Subjects and Methods section.
(E) Immunoblot analysis of PHF21A levels in DGAP012 and MCN1762 and in controls. The PHF21A antibody against an N-terminal 93 residue polypeptide28 recognizes the ∼92 kDa PHF21A in both female (“F”) and male (“M”) controls, as well as in DGAP012 and MCN1762 lymphoblastoid cell line extracts. It shows notably reduced protein levels due to disruption of PHF21A (arrow) in both DGAP012 and MCN1762. A 73 kDa protein (arrowhead) was noted in DGAP012 and is likely to be a product of the PHF21AΔex15-18/ELAVL1Δex1-5 fusion gene, which deletes the critical plant homeodomain (PHD) finger domain. α-tubulin was used as an internal loading control. The bar graph shows the mean and ± standard deviations from three independent experiments (∗p < 0.001, ∗∗p < 0.0001). The subcellular localization of PHF21A with the same antibody is described in Figure S3.
(F) PHF21A functional domains in wild-type and theoretical truncated proteins in two balanced translocation subjects. PHF21A contains two leucine zipper domains (LZD1 and LZD2), one AT-hook domain, and one PHD zinc finger domain. The amino acid positions of all domains are indicated as numbers below the domain structures. Note that if any protein were produced from the truncated PHF21A of DGAP012 or MCN1762, it would lack the PHD finger domain essential for binding H3K4me0.14
