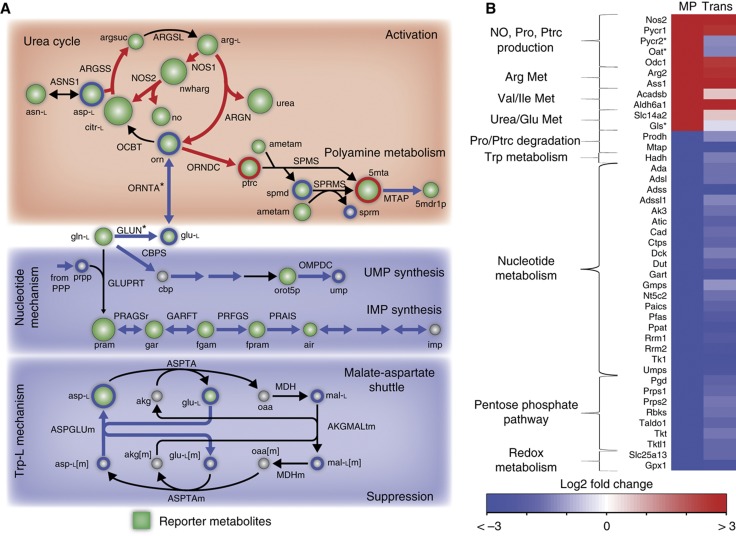
Figure 4.
High-throughput data support in-silico predictions. (A) Reporter metabolites provide a global analysis of the expression data. Major changes pertained to predicted pathways of activation and suppression. Green nodes are scaled by degree of enrichment. Circled metabolites in red and blue represent significantly changed metabolites detected by GC–MS. (B) Directionality of in-silico predictions was in high accordance with the transcriptional and proteomic response of LPS-stimulated cells. Pycr2, Oat, and Gls expression contradicted model predictions, but the proteomics data confirmed the predictions. Only 24 h transcriptomics data are shown due to sparsity of proteomic data. MP – Model Prediction, metabolite, and reaction abbreviations are provided in Supplementary information.