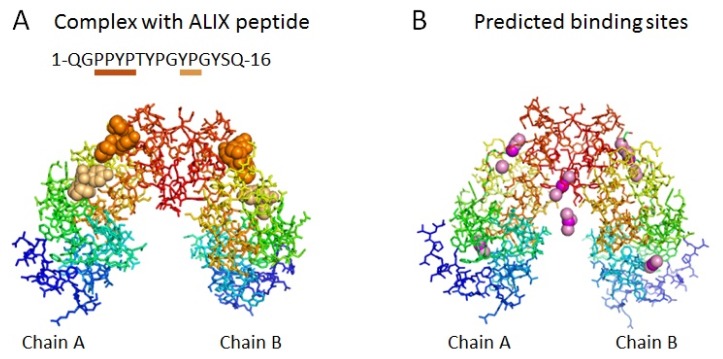
Figure 1.
Ligand-binding sites in ALG-2 dimer. (A) Crystal structure of the Zn2+-bound form of des3-23ALG-2 in the complex with ALIX ABS peptide (PDB ID: 2zne). ALG-2 dimer molecules (chains A and B) are drawn by PyMol and displayed by stick models in rainbow colors (blue to red gradation from N-terminal to C-terminal regions). Orange and light orange spheres indicate PPYP and YP moieties of the ALIX peptide (underlined) bound to ALG-2. The ALIX peptide bound to chain B is shown for YP from chain D and for PPYP from chain H in two-fold crystallographically symmetric ALG-2 tetramer (chains A, B, E and F) that is in complex with four peptide molecules with two different binding patterns (chains C, D, G and H. see Supplementary file of 2zne_and_symmetry in PDB format). Zinc atoms are not shown. (B) Top six ranked ligand-binding sites predicted in the ALG-2 dimer (des3-20ALG-2, PDB ID: 2zn9) by MetaPocket 2.0 using chains A and B of PDB ID 2zn9. Spheres colored in magenta and light pink indicate predicted centers of ligand-binding sites by MetaPocket and top three sites by individual predictors, respectively. Calcium atoms are not shown.