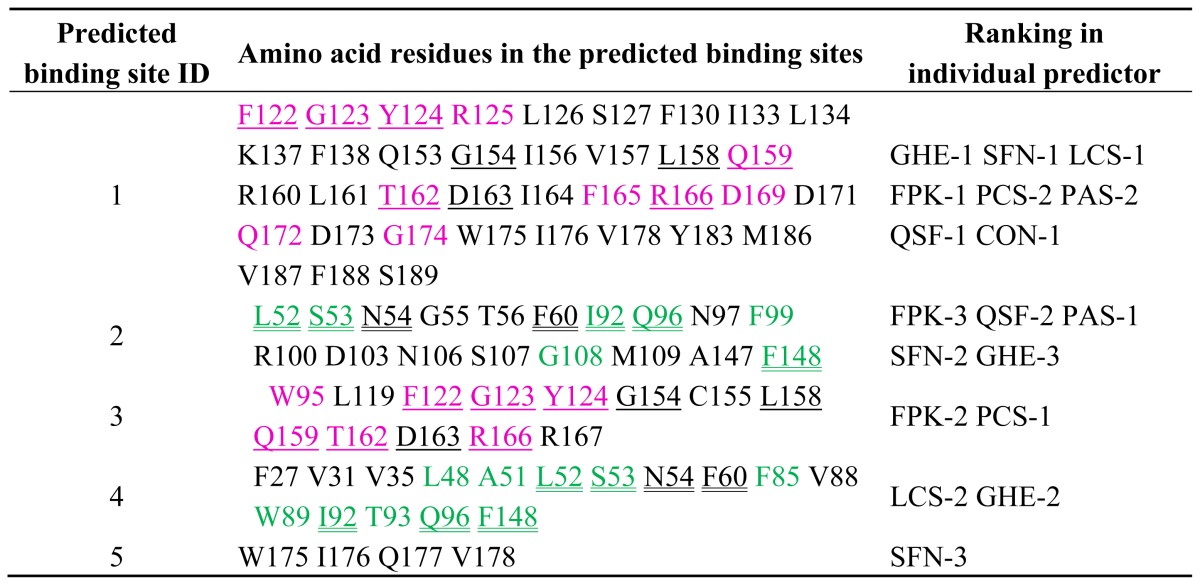
Table 1.
Potential ligand-binding sites in the monomer molecule of ALG-2 predicted by multiple computational algorithms. Chain A of PDB ID 2zn9 was searched for potential ligand-binding sites using MetaPocket 2.0 that combines the results of eight predicting methods. Predicted top five ranked binding sites (ID Nos. 1–5) and the residue numbers of amino acids that form the potential binding sites are listed. Letters in magenta in binding site ID Nos. 1 and 3 are residues known to interact with the ALIX peptide in the crystal structure (PDB ID 2zne, chains A and C). Letters in green in binding sites ID Nos. 2 and 4 are residues predicted to interact with a model type 2 motif peptide by simulation with AutoDock Vina and analyzed with LIGPLOT (See Table 3). Each residue overlapping between ID Nos. 1 and 3 and between ID Nos. 2 and 4 is indicated by an underline and double underline, respectively. Number following the hyphen indicates ranking in individual predictors used in MetaPocket 2.0: ConCavity (CON); Fpocket (FPK); GHECOM (GHE); LigsiteCS (LCS); PASS11 (PAS); POCASA (PCS); Q_SiteFinder (QSF); SURFNET (SFN).