Background: α4 binds to the PP2A catalytic subunit and the microtubule-associated E3 ligase MID1.
Results: MID1-dependent monoubiquitination promotes calpain-mediated cleavage of α4, altering its phosphatase regulatory function.
Conclusion: Defects in this regulatory process may underlie the MAP hypophosphorylation and hyperphosphorylation seen in Opitz syndrome and Alzheimer disease.
Significance: Pharmacological agents that interfere with α4 monoubiquitination or cleavage are potential therapeutics to treat Alzheimer disease.
Keywords: Alzheimer Disease, E3 Ubiquitin Ligase, Phosphatase, PP2A, Ubiquitination, alpha4, Microtubule-associated Protein
Abstract
Multiple neurodegenerative disorders are linked to aberrant phosphorylation of microtubule-associated proteins (MAPs). Protein phosphatase 2A (PP2A) is the major MAP phosphatase; however, little is known about its regulation at microtubules. α4 binds the PP2A catalytic subunit (PP2Ac) and the microtubule-associated E3 ubiquitin ligase MID1, and through unknown mechanisms can both reduce and enhance PP2Ac stability. We show MID1-dependent monoubiquitination of α4 triggers calpain-mediated cleavage and switches α4's activity from protective to destructive, resulting in increased Tau phosphorylation. This regulatory mechanism appears important in MAP-dependent pathologies as levels of cleaved α4 are decreased in Opitz syndrome and increased in Alzheimer disease, disorders characterized by MAP hypophosphorylation and hyperphosphorylation, respectively. These findings indicate that regulated inter-domain cleavage controls the dual functions of α4, and dysregulation of α4 cleavage may contribute to Opitz syndrome and Alzheimer disease.
Introduction
Deregulation of protein phosphatase 2A (PP2A)2 has been implicated in a variety of microtubule-associated protein (MAP)-dependent pathologies such as Alzheimer disease (AD), Opitz syndrome (OS), and various cancers (1–3). Several regulatory mechanisms have been described for PP2A, including post-translational modifications of the PP2A catalytic subunit (PP2Ac) and the association of PP2Ac with regulatory subunits (4, 5). Although the most studied forms of PP2A are the heterotrimeric holoenzymes, which consist of PP2Ac, a structural A subunit and a variable B subunit that dictates substrate selectivity and subcellular localization of the phosphatase holoenzyme, PP2Ac also interacts with a number of atypical regulatory subunits independent of the canonical A and B subunits. Within this group is α4, a direct interacting partner of PP2Ac that also binds the microtubule-associated E3 ubiquitin ligase MID1 and plays a crucial role in modulating PP2Ac polyubiquitination and stability (6–9). Loss of function mutations in the MID1 gene are the underlying cause of OS (10), a congenital disorder characterized by defects in midline development and significant increases in microtubule-associated PP2A activity (1). α4 initially was shown to tether PP2Ac to MID1 and promote polyubiquitination and degradation of microtubule-associated PP2Ac (1); however, subsequent studies revealed that α4 can also protect PP2Ac from polyubiquitination and proteasomal degradation (7–9). These findings indicate that α4 may exhibit both protective and destructive actions in the control of PP2Ac cellular levels.
In this report, we demonstrate that MID1 functions as the E3 ubiquitin ligase for monoubiquitination of α4, which triggers calpain-mediated cleavage of the C-terminal MID1-binding domain of α4. Furthermore, we demonstrate that PP2Ac stability is influenced by the monoubiquitination state of α4. Monoubiquitination of α4 promotes calpain-mediated cleavage and switches its activity from protective to destructive toward PP2Ac. Consistent with these results, we observed increased phosphorylation of the microtubule-associated Tau protein in cells overexpressing the N-terminal cleavage product of α4 relative to cells overexpressing full-length α4. Finally, analysis of post-mortem AD tissue and cells derived from an OS fetus revealed marked alterations in α4 cleavage, thus indicating that defective α4 monoubiquitination/cleavage and consequential deregulation of PP2A function may be involved in the pathogenesis of OS and AD.
EXPERIMENTAL PROCEDURES
Human Tissue
After receiving human subjects approval from the University of Washington human subjects division, we obtained de-identified samples of post-mortem temporal cortex tissue from AD and age-matched control cases from the University of Washington Alzheimer Disease Research Center Neuropathology Core (Core leader, Dr. Thomas Montine). The tissues were lysed by sonication in a high salt buffer, and the clarified lysates were analyzed via Western blot.
Cell Culture and Transfections
HEK293FT cells were cultured in DMEM supplemented with 10% fetal calf serum and 2 mm l-glutamine. HEK/Tau stable cells (11) were grown under the same conditions except the media also contained 100 μg/ml Zeocin to maintain Tau selection. Human embryonic fibroblasts (HEFs) derived from a fetus with Opitz syndrome and from an age-matched control fetus were described previously (15) and grown in DMEM supplemented with 10% fetal calf serum and 2 mm l-glutamine. HEK293FT and HEK/Tau cells were transfected with mammalian expression constructs using FuGENE 6 (Roche Applied Science) according to the manufacturer's protocol. MID1-targeted siRNA was introduced into HEK293FT cells using Dharmafect (Thermo Fisher Scientific) according to the manufacturer's protocol.
In Vitro Ubiquitination Assays
The ability of MID1·E2 pairs to form polyubiquitin chains was determined using the E3LITE customizable ubiquitin ligase kit (LifeSensors). PP2Ac and α4 ubiquitination assays were performed using either a ubiquitin-protein conjugation kit (BostonBiochem) as described previously (9) or purified conjugation enzymes. Details of the in vitro ubiquitination assays can be found in the supplemental “Materials and Methods.”
Mass Spectrometry (MS)
FLAG-α4 was immunopurified from HEK293FT cells and subjected to SDS-PAGE. Full-length FLAG-α4 and the proteolytic fragment of FLAG-α4 were visualized by Colloidal Blue staining and excised from the gel. Excised gel fragments were digested with trypsin and analyzed by C18 reverse-phase LC-MS/MS using a Thermo LTQ-XL Orbitrap ion trap tandem mass spectrometer. Tandem MS data were analyzed with the Sequest algorithm.
Cycloheximide Chase Experiments
HEK293FT cells transfected with either HA3-PP2Ac alone or together with FLAG-α4 WT, FLAG-α4 ΔUIM, or FLAG-α4 A52L/A53L were treated with 100 μg/ml cyclohexamide (Sigma) for the indicated times. Cell lysates were then prepared and subjected to Western analysis.
Immunoprecipitations
Clarified lysates from HEK293FT cells expressing FLAG- or HA-tagged proteins were incubated with anti-HA-agarose (Roche Applied Science) or anti-FLAG-agarose (Sigma). Bound proteins were washed, eluted in SDS sample buffer, and subjected to Western analysis. In some cases, the FLAG-tagged proteins were eluted from the beads by incubation in buffer containing 100 μg/ml FLAG peptide.
In Cell Western Analysis
HEK293/Tau cells expressing FLAG-α4 wild type or FLAG-α4 Gly256* were fixed in a 96-well plate with 4% paraformaldehyde and then permeabilized in 1× PBS containing 0.1% Triton X-100. The permeabilized cells were incubated with primary antibodies (diluted 1:1000 in Odyssey blocking buffer) overnight at 4 °C and incubated with secondary antibodies (diluted 1:500 in Odyssey blocking buffer containing 0.2% Tween 20) for 60 min at room temperature. Bound antibodies were visualized and quantified using the Odyssey Infrared Imaging system and Odyssey software.
Statistics
Data are expressed as mean ± S.E. Statistical comparisons were performed using an unpaired Student's t test and analysis of variance when appropriate.
Additional experimental details, as well as a list of plasmids and antibodies, can be found in the supplemental “Materials and Methods.”
RESULTS
MID1 Is E3 Ubiquitin Ligase for α4
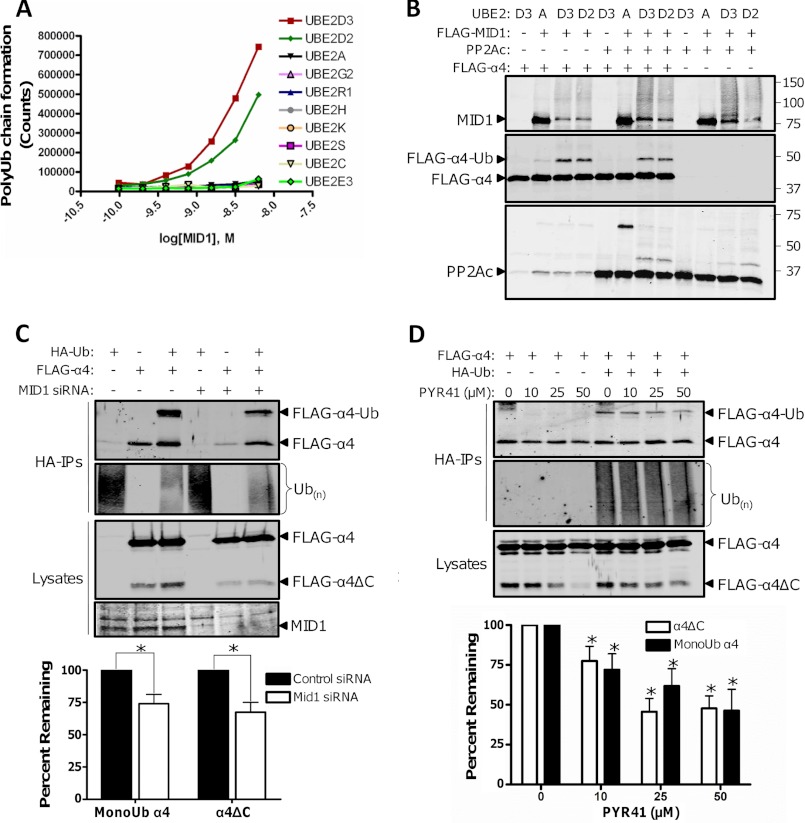
MID1, α4, and PP2Ac form a ternary complex in cells (9). Both α4 and PP2Ac are targeted for ubiquitination (monoubiquitination of α4 and polyubiquitination of PP2Ac) (1, 9, 12), but the target of MID1 remains unclear. Although MID1 was initially postulated to function as the E3 ubiquitin ligase for PP2Ac polyubiquitination (1), a recent in vitro study demonstrated that the RING and B-box domains of MID1 possess E3 ligase activity and can monoubiquitinate a 45-amino acid polypeptide derived from the C terminus of α4 (12). However, no reports have examined whether full-length MID1 directly promotes PP2Ac and/or α4 ubiquitination. To determine whether full-length MID1 possesses E3 ligase activity and to identify the E2 ubiquitin-conjugation enzymes that pair with MID1, we assayed the ability of immunopurified full-length MID1 to form polyubiquitin chains using a panel of purified E2 enzymes. As shown in Fig. 1A, a MID1 dose-dependent increase in polyubiquitin chain formation was observed in presence of the E2 enzymes UBE2D3 and UBE2D2, but no detectable polyubiquitin chain formation was seen in reactions containing the other E2 enzymes. To further characterize the ligase activity of MID1, we incubated FLAG-MID1 with α4 and/or PP2Ac in ubiquitination assay mixtures containing purified E1 and various E2 enzymes. The MID1-UBE2D3 and MID1-UBE2D2 pairs, but not the MID1-UBE2A pair, facilitated α4 monoubiquitination and MID1 auto-polyubiquitination in the presence or absence of PP2Ac (Fig. 1B). No appreciable PP2Ac polyubiquitination was observed in any of the experimental conditions; however, a faint signal corresponding to monoubiquitinated PP2Ac was observed in the reactions containing the MID1-UBE2D3 and MID1-UBE2D2 pairs. Because recent reports have shown that some E3 ubiquitin ligases require multiple E2 enzymes to facilitate protein polyubiquitination (13, 14), we tested whether multiple E2 enzymes are necessary for MID1-dependent PP2Ac polyubiquitination. Purified PP2Ac was incubated in a ubiquitin-protein conjugation solution containing the E1 enzyme and a mixture of E2 enzymes (fraction A), and either a mixture of E3 ligases (fraction B) or immunopurified FLAG-MID1. Western analysis of the reactions containing the E3 ligase mixture revealed polyubiquitinated PP2Ac species, whereas analysis of reactions containing FLAG-MID1 showed no detectable polyubiquitinated PP2Ac species (supplemental Fig. S1). Together, these data indicate that MID1 functions as the E3 ubiquitin ligase for α4 monoubiquitination but fails to promote PP2Ac polyubiquitinaton even in the presence of multiple E2 enzymes. Moreover, these data raise the possibility that a different E3 ligase is responsible for PP2Ac polyubiquitination.
FIGURE 1.
MID1-dependent monoubiquitination promotes cleavage of the C terminus of α4. A, the indicated E2 enzyme was incubated with increasing concentrations of purified FLAG-MID1for 1 h, and the ability of each MID1/E2 pair to form polyubiquitin chains was measured using the customizable E3LITE ubiquitin ligase kit. B, ubiquitination assays were performed for 1 h in the presence (+) or absence (−) of purified FLAG-MID1, PP2Ac, FLAG-α4, and the indicated E2 enzyme (UBE2). Ubiquitinated MID1, α4, and PP2Ac species were visualized by Western analysis using the corresponding antibodies. C, HEK293FT cells were transfected with HA-ubiquitin (HA-Ub) and FLAG-α4 plasmids alone or together with MID1 siRNA (+) or control siRNA (−). Ubiquitinated proteins were isolated from the cell extracts using a HA-affinity matrix (HA-IPs). The HA-IPs and cell lysates were analyzed by Western using antibodies recognizing α4 and ubiquitin. The cell lysates were also analyzed by Western using a MID1-specific antibody to confirm protein knockdown. The monoubiquitinated α4 and cleaved α4 signals were normalized to the total α4 signal in the corresponding cell lysate, and the normalized monoubiquitinated and cleaved α4 values from the MID1 knockdown conditions were compared with the corresponding values in the control siRNA conditions, which were set at 100. Values represent means ± S.E. *, p < 0.001. Some unmodified FLAG-α4 (nonspecific) was detected in the HA-IPs, but the levels of unmodified FLAG-α4 were not statistically different between the control and experimental condition from multiple experiments. D, HEK293FT cell expressing HA-Ub and/or FLAG-α4 were treated with increasing concentrations of PYR41 for 4 h prior to lysis. Monoubiquitinated FLAG-α4 and total ubiquitinated proteins were visualized by Western analysis of the HA-IPs using α4- and ubiquitin-specific antibodies. Cell lysates were also subjected to Western analysis using an α4 antibody. The α4 monoubiquitin (MonoUb) and α4 cleavage signals from PYR41-treated cells were normalized to the signal from untreated cells. Values represent means ± S.E. *, p < 0.01.
To determine whether MID1 facilitates α4 monoubiquitination in mammalian cells, we co-transfected HEK293FT cells with HA-ubiquitin and FLAG-α4, together with control siRNA or MID1-targeted siRNA. The expression of endogenous MID1 was reduced dramatically in cells transfected with MID1 siRNA relative to cells transfected with control siRNA (Fig. 1C). Western analysis of the ubiquitinated proteins isolated from the cell lysates revealed significantly decreased α4 monoubiquitination but not total protein ubiquitination in cells harboring MID1 siRNA compared with the control cells (Fig. 1C). These cellular findings further establish MID1 as the E3 ubiquitin ligase for α4.
Monoubiquitination of α4 Promotes Its Cleavage
α4 is a multi-domain protein with an unstructured C terminus (8, 15, 16). We detected a proteolytic fragment of α4 (27 kDa) in lysates of cells expressing an N-terminal FLAG-tagged form of this protein (Fig. 1C). Both full-length and truncated α4 were immunoreactive with a FLAG antibody and an antibody directed against the N terminus of α4, but only the full-length protein was recognized with an antibody directed against the C terminus of α4 (Fig. 1C and data not shown), thus demonstrating that in cells α4 is subject to proteolytic cleavage resulting in a truncated protein that lacks the C terminus (α4ΔC). Unexpectedly, we also observed that the amount of α4 cleavage product appeared to parallel the monoubiquitination state of α4 as cells harboring MID1 siRNA exhibited decreased cleavage and monoubiquitination in comparison with control cells (Fig. 1C). Quantification of these data revealed that the MID1 siRNA-induced decrease in α4 cleavage and monoubiquitination was not due to alterations in total FLAG-α4 levels (Fig. 1C, bottom panel). These findings point to an unprecedented monoubiquitination-regulated proteolysis event.
To better understand the relationship between α4 monoubiquitination and cleavage and to rule out any MID1-independent effects of the MID1 siRNA on α4 cleavage, we treated target cells with increasing concentrations of the E1 enzyme inhibitor PYR41. As shown in Fig. 1D, the monoubiquitination and cleavage of α4 both decreased in a strikingly similar PYR41 dose-dependent manner. In agreement with previous reports (17), we did not observe significant changes in total ubiquitin conjugates with PYR41 (Fig. 1D). These data further illustrate that alterations in α4 monoubiquitination influence its own cleavage.
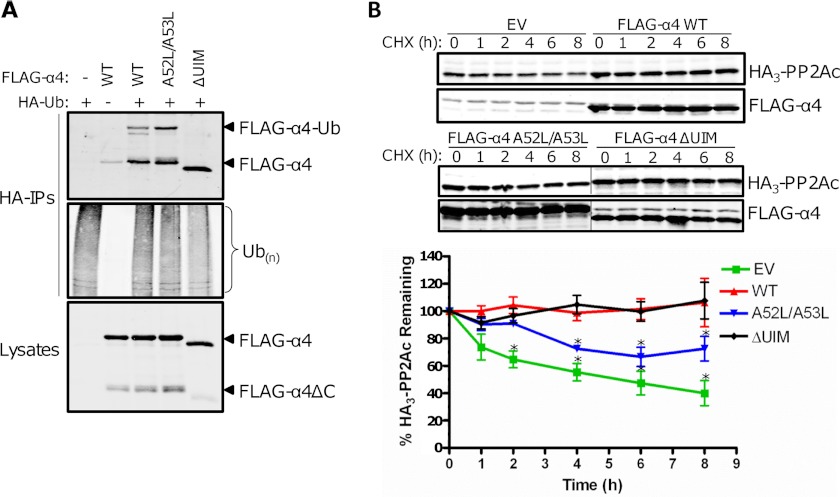
Because our previous study demonstrated that human α4 contains a ubiquitin-interacting motif (UIM; residues 46–60) (9), we asked whether mutation or deletion of this motif influenced α4 monoubiquitination and cleavage product formation. Like other UIM-containing containing proteins (18), we found that deletion of the UIM within α4 (ΔUIM) prevented the protein from undergoing monoubiquitination (Fig. 2A). Our analyses of multiple α4 UIM point mutants also identified a double point mutant of α4 (A52L/A53L) that exhibited increased monoubiquitination (Fig. 2A). Importantly, notable differences in the amount of cleavage product (α4ΔC) were observed in these cells; A52L/A53L-expressing cells exhibited higher levels of α4ΔC relative to wild type α4-expressing cells, but very little α4ΔC was detected in ΔUIM-expressing cells (Fig. 2A). Thus, α4 cleavage is dependent on its monoubiquitination: A52L/A53L > wild type α4 ≫ ΔUIM. These findings, together with the results of the MID1 knockdown and PYR41 experiments, support our hypothesis that MID1-dependent monoubiquitination of α4 triggers its proteolytic cleavage.
FIGURE 2.
α4 monoubiquitination and cleavage are essential for PP2Ac turnover. A, HEK293FT cells were transfected with HA-ubiquitin (HA-Ub) and either empty vector (−), wild type FLAG-α4 (WT), a UIM deletion mutant of FLAG-α4 (ΔUIM), or a FLAG-α4 double point mutant (A52L/A53L). Western analysis of the HA-IPs was performed using α4 and ubiquitin antibodies. Cell lysates were similarly probed using an α4-specific antibody. B, HEK293FT cells expressing HA3-PP2Ac alone or together with empty vector (EV), FLAG-α4 WT, A52L/A53L, or ΔUIM were treated with 100 μg/ml cycloheximide (CHX) at 48 h post-transfection and then lysed at the indicated time points after treatment. The lysates were analyzed by Western using antibodies recognizing HA3-PP2Ac and α4. Samples were analyzed for statistically significant changes relative to the corresponding HA3-PP2Ac + FLAG-α4 WT samples. *, p < 0.05 (analysis of variance).
α4 Monoubiquitination and Cleavage Are Important in Regulation of PP2Ac Stability
The gene encoding α4 (IGBP1) is an essential gene as its deletion causes lethality of the host and cellular apoptosis (19). Furthermore, studies of conditional α4-null mouse embryonic fibroblasts have revealed that α4 plays a crucial role in the maintenance of PP2Ac stability (7). To explore a potential role of α4 monoubiquitination and cleavage in the regulation of PP2Ac stability, we performed cycloheximide chase studies of cells expressing HA3-PP2Ac alone or together with various FLAG-tagged α4 constructs exhibiting differing degrees of monoubiquitination and cleavage. The cells were treated with cycloheximide 48 h post-transfection to inhibit new protein synthesis, and the levels of HA3-PP2Ac were monitored at various time points after cycloheximide treatment (Fig. 2B). Consistent with our previous report (8), we observed a progressive decline in HA3-PP2Ac levels over the 8 h cycloheximide time course in cells expressing HA3-PP2Ac alone, but the levels of HA3-PP2Ac remained stable during this period in cells coexpressing wild type α4. Surprisingly, HA3-PP2Ac levels also remained stable in cells coexpressing ΔUIM after cycloheximide treatment; however, the levels of HA3-PP2Ac progressively declined in cells coexpressing A52L/A53L. These results indicate that the monoubiquitination- and cleavage-resistant ΔUIM construct prevents HA3-PP2Ac turnover, whereas the A52L/A53L mutant, which exhibits increased monoubiquitination and cleavage, allows HA3-PP2Ac turnover. Furthermore, these data highlight a role for α4 monoubiquitination and cleavage in the control of PP2Ac turnover.
Calpain-mediated Cleavage of α4 Occurs at Phe255-Gly256 Bond
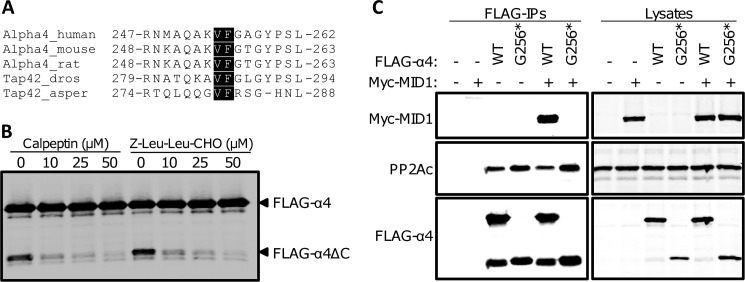
We next sought to determine the cleavage site in α4. Electrospray ionization high-pressure liquid chromatography tandem mass spectrometry of α4 and α4ΔC identified multiple overlapping peptides that corresponded to the N-terminal portion of α4. However, one peptide was found to be unique to α4ΔC, NMAQAKVF (supplemental Fig. S2); no α4 peptides beyond this region were identified in the cleaved protein, but peptides covering the entire sequence were identified in the full-length α4 sample (Fig. 3A). Western analysis of cells expressing FLAG-tagged wild type α4 or an α4 construct encompassing residues 1–255 (Gly256*) revealed that Gly256* comigrated exactly with the authentic cleavage fragment derived from full-length FLAG-α4 (Fig. 3B). Together, these data demonstrate that the 27-kDa fragment of α4 (α4ΔC) is the result of proteolytic cleavage of the full-length protein between residues Phe255 and Gly256.
FIGURE 3.
α4 cleavage occurs at the Phe255-Gly256 bond. A, coverage map of the full-length α4 and cleaved α4 peptides determined by mass spectrometry. Closed boxes mark peptide regions identified in full-length FLAG-α4; open boxes mark peptide regions identified in the FLAG-α4 cleavage product. The residues highlighted in gray correspond to the unique peptide found only in the α4 cleavage product. The MS spectrum for this peptide can be found in supplemental Fig. S2. B, HEK293FT cells expressing empty vector (EV), wild type FLAG-α4 (WT), or FLAG-α4 1–255 (G256*) were analyzed by Western using an α4-specific antibody.
Analysis of the amino acid residues flanking the α4 cleavage site identified a potential calpain consensus sequence (Fig. 4A). To test whether calpains are responsible for the cleavage of α4, we treated cells expressing FLAG-α4 with increasing concentrations of the calpain inhibitors calpeptin or Z-Leu-Leu-CHO and monitored cleavage product formation by Western analysis. The calpain inhibitors potently protected FLAG-α4 from cleavage (Fig. 4B). Because the VF residues are conserved among many different species of α4 (Fig. 4A), calpain-mediated cleavage at Phe255 may represent an evolutionarily conserved mode of regulation for α4. Additional support for this proposal comes from our observations showing that the Drosophila homolog of α4, Tap42, is also targeted for both monoubiquitination and cleavage (supplemental Fig. S3).
FIGURE 4.
Calpain-mediated cleavage of α4 results in an N-terminal fragment that binds PP2Ac but not MID1. A, sequence alignment of the region within multiple species of α4 that encompasses the calpain cleavage site; the conserved VF residues are highlighted. B, HEK293FT cells expressing wild type FLAG-α4 were treated with increasing concentrations of calpeptin or Z-Leu-Leu-CHO for 4 h prior to lysis. Cell lysates were subjected to Western analysis using an α4 antibody. C, HEK293FT cells were transfected with wild type FLAG-α4 WT and Gly256* alone or together with (+) or without (−) Myc-MID1. FLAG-tagged proteins were immunoprecipitated (FLAG-IPs) from the cell lysates and analyzed by Western using the Myc, PP2Ac, and α4 antibodies. The cell lysates were analyzed similarly. asper, Aspergillus; dros, Drosophila.
Cleavage of α4 Regulates Its Interaction with MID1
Previous studies have shown that PP2Ac binds to the N-terminal domain of α4 (16), whereas MID1 binds to the C-terminal domain of α4 (1, 6), yet both domains are required for α4-mediated protection of PP2Ac from polyubiquitination and degradation (8). Because α4 cleavage (at the Phe255-Gly256 bond) occurs within the previously identified MID1 binding region, we performed experiments to determine whether the cleavage product of α4 still retains the ability to bind MID1. Western analysis of FLAG immune complexes from HEK293FT cells co-expressing Myc-MID1 and either wild type FLAG-α4 or FLAG-α4 Gly256* revealed that both forms of α4 bound to PP2Ac, but only full-length α4 interacted with MID1 (Fig. 4C). These results demonstrate that the C-terminal 84 amino acids of human α4 (amino acids 256–340) are necessary for MID1 binding. Furthermore, these findings indicate that α4 cleavage likely leads to the disruption of MID1·α4·PP2Ac complexes.
Regulation of Tau Phosphorylation by α4 Cleavage
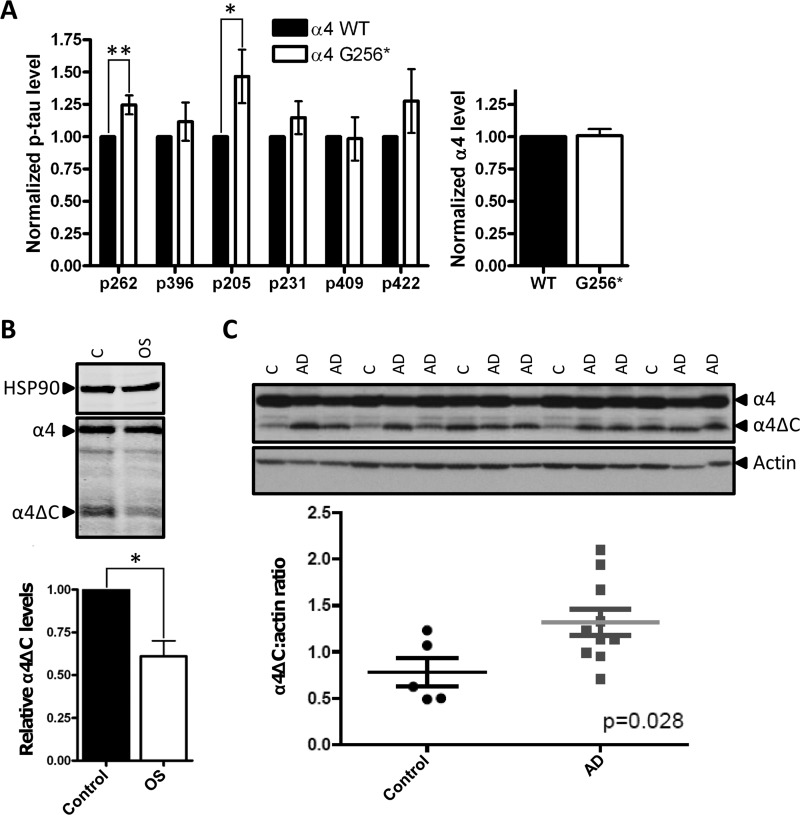
The MID1·α4·PP2A complex is localized to microtubules via the interaction of MID1 with microtubule structures and is thought to be involved in the maintenance of microtubule stability (6, 20, 21). Although the precise function of this complex in microtubule stabilization remains unclear, it likely involves PP2A-mediated dephosphorylation of various MAPs. Tau is one such MAP that is targeted for PP2A dephosphorylation at multiple epitopes (22). To test whether α4 cleavage plays a role in the control of PP2Ac-dependent Tau dephosphorylation, we transfected HEK293 cells stably expressing the Tau protein (11) with HA3-PP2Ac and either wild type FLAG-α4 or FLAG-α4 Gly256*. The levels of Tau Ser262 and Thr205 phosphorylation were significantly elevated in FLAG-α4 Gly256*-expressing cells relative to wild type FLAG-α4-expressing cells as monitored by in-cell Western blots (Fig. 5A). These data support the hypothesis that α4 cleavage leads to a loss of PP2Ac at microtubules and consequential increased phosphorylation of PP2A-sensitive sites within Tau.
FIGURE 5.
Cleaved α4 increases Tau phosphorylation and is altered in OS and AD. A, HEK/Tau cells expressing HA3-PP2Ac and either wild type FLAG-α4 (α4 WT) or FLAG-α4 Gly256* (α4-G256*) were fixed in a 96-well plate and subjected to in-cell Western blotting using antibodies recognizing the FLAG epitope, HSP90, total Tau, or the indicated phospho-Tau epitope. The phospho-Tau signal was normalized to the total Tau signal in each well, and the normalized phospho-Tau values in α4 Gly256*-expressing cells were compared with the corresponding values in α4 WT-expressing cells, which were set at 1. Likewise, the FLAG-α4 signal was normalized to the HSP90, and the normalized α4 values in Gly256*-expressing cells were compared with corresponding values in α4 WT-expressing cells, which were set at 1. Values represent means ± S.E. *, p < 0.05, **, p < 0.01. B, lysates from Opitz syndrome-derived HEFs and aged-matched control HEFs were analyzed by Western blotting using antibodies recognizing HSP90, α4, and PP2Ac. The graph depicts the relative α4ΔC levels (ratio of α4ΔC signal to total α4 signal (α4ΔC + full-length α4)) in the two cell types with the relative α4ΔC levels in control HEFs set at 1. Values represent means ± S.E. *, p < 0.001 from three independent experiments using the two cell lines. C, trios of age-matched AD cases (n = 10) and control patients (n = 5) post-mortem temporal cortex tissue samples were analyzed by Western for full-length α4 and α4ΔC levels as described above. Samples are loaded in order of descending age. AD represents an Alzheimer disease case, C is a normal control case. Quantification of α4ΔC levels were carried out using Adobe Photoshop analysis functions and normalized to actin levels. Mean α4ΔC/actin ratios are 1.3 ± 0.14 for AD cases and 0.78 ± 0.15 for controls. Depicted error represents S.E. The difference between AD cases and controls is statistically significant; p = 0.028.
α4 Cleavage Is Decreased in OS
Loss of function mutations in the MID1 gene are the underlying cause of OS (10), a congenital disorder characterized by defects in midline development, increases in microtubule-associated PP2A activity, and a global hypophosphorylation of MAPs (1). To determine whether α4 cleavage is altered in OS, we probed lysates of human embryonic fibroblasts derived from a fetus with OS (OS-HEFs) and control HEFs for the presence of the α4 cleavage product (α4ΔC). The amount of cleaved α4 detected in the OS-HEFs was significantly reduced compared with control HEFs (Fig. 5B), thus supporting a potential role for deregulated α4 cleavage in the pathogenesis of OS. Furthermore, like the HEK293FT cells expressing FLAG-α4 (Fig. 4B), treatment of the HEFs with calpain inhibitors also protected endogenous α4 from cleavage (data not shown). These findings support the notion that defects in MID1 function, as is the case in OS cells, may lead to reduced α4 monoubiquitination/cleavage and increased PP2A levels at microtubules, which may explain the hypophosphorylation of MAPs seen in OS (1).
α4 Cleavage Is Increased in AD
Amyloid-β-containing plaques and neurofibrillary tangles, composed of hyperphosphorylated forms of Tau, are hallmark features of AD (23). Although the precise role of amyloid-β plaques and neurofibrillary tangles in disease progression remains unknown, recent evidence indicates that these lesions are closely linked and points to a role for abnormally hyperphosphorylated Tau in amyloid-β toxicity (24, 25). To determine whether deregulation of Tau during disease coincides with alterations in α4 cleavage and hence PP2Ac stability, we analyzed α4 cleavage in temporal cortex tissue samples from 10 Alzheimer cases and five control cases. As shown in Fig. 5C, α4 cleavage was significantly enhanced in AD brains.
DISCUSSION
In this report, we show that MID1 possesses E3 ligase activity and directly facilitates α4 monoubiquitination, which triggers calpain-mediated cleavage of the PP2A regulatory subunit. Monoubiquitination has been shown previously to impact the activity and subcellular localization of many proteins (26); however, to our knowledge, monoubiquitination-induced cleavage represents a hitherto undescribed activity for ubiquitin. Considering the relatively large number of cellular proteins that undergo both monoubiquitination and cleavage (e.g. IL-1R1 (27) and Notch (28)), it will be interesting to determine whether monoubiquitination-induced cleavage represent a more general phenomenon for the control of protein function.
How is the monoubiquitinated form of α4 targeted for cleavage? Deletion of the UIM within α4 prevents its monoubiquitination and cleavage, thus supporting the idea that a functional UIM is necessary for these events. We propose that the UIM within α4, which is known to form non-covalent interactions with ubiquitin (9), binds in cis to the ubiquitin moiety on α4 leading to a conformational change in α4 that unmasks a calpain cleavage site. Although we speculate a cis conformational change, we cannot rule out the possibility that the conformational change could be occurring in trans. Studies of calpain substrates have revealed that the tertiary structure of the protein is important for cleavage (29); therefore, a monoubiquitin-induced conformational change in α4 could explain why only a fraction of α4 (i.e. monoubiquitinated α4) is targeted for cleavage.
Our studies demonstrate that α4 monoubiquitination and cleavage are important for facilitating PP2Ac degradation. Because the cleaved form of α4 (α4ΔC) retains its ability to bind PP2Ac but fails to bind MID1, α4 cleavage could be important for redirecting the localization of PP2Ac and promoting the polyubiquitination of this phosphatase by a yet unknown E3 ubiquitin ligase. Although the ubiquitination machinery necessary for PP2Ac ubiquitination remains to be identified, our studies provide compelling support for new model of MID1/α4 regulation of PP2Ac in which MID1-mediated monoubiquitination of α4 triggers a conformational change in α4 leading to calpain-cleavage of its MID1 binding domain and, ultimately, PP2Ac polyubiquitination and proteasomal degradation (Fig. 6). Our studies also reconcile the apparent contradictory protective and destructive roles of α4 in the control of PP2Ac levels (1, 7–9) and place α4 in a critical position where it can signal either protection or degradation of PP2Ac depending on the ubiquitination state of α4. The cytosolic form of α4 that is not associated with MID1 likely stabilizes PP2Ac in an inactive form until it can be incorporated into active PP2A holoenzymes (7), whereas the microtubule-associated MID1·α4-bound PP2Ac is subject to proteasomal degradation as a result of MID1-dependent monoubiquitination and cleavage of α4.
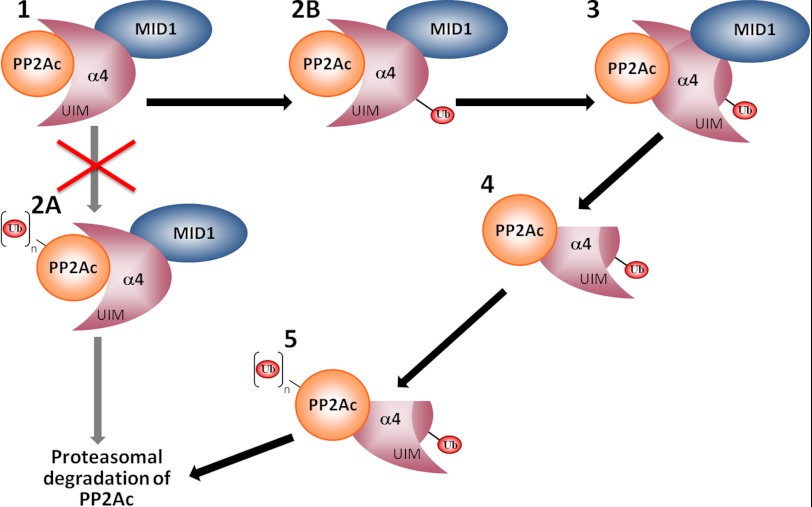
FIGURE 6.
Working model for MID1/α4 regulation of PP2Ac. MID1, α4, and PP2Ac form a ternary complex (1) in cells. Previous studies have suggested that α4 facilitates MID1-dependent polyubiquitination of PP2Ac (2A) and subsequent proteasomal degradation; however, our findings challenge this paradigm and support a model in which MID1 serves as the E3 ligase for α4 (2B), leading to a conformational change in α4 whereby the UIM of α4 binds in cis to the covalently attached ubiquitin (Ub; 3). This structural rearrangement then leads to calpain-mediated cleavage of the C terminus of α4 (4), allowing for polyubiquitination of PP2Ac by a currently unknown E3 ligase (5) and subsequent degradation by the proteasome.
Our studies indicate that MID1-dependent regulation of PP2Ac levels at microtubules (via α4 monoubiquitination/cleavage-induced PP2Ac degradation) plays a crucial role in maintaining the proper phosphorylation state of MAPs. Defects in this phosphatase regulatory process such as those that occur in OS (mutated MID1, decreased α4 cleavage, stabilization of microtubule-associated PP2Ac) and AD (increased α4 cleavage, destabilization of microtubule-associated PP2Ac) may explain the MAP hypophosphorylation and Tau hyperphosphorylation seen in OS and AD, respectively. Previous reports have shown that the ABαC holoenzyme is the major form of PP2A responsible for Tau dephosphorylation (30, 31) and that the A subunit competes with α4 for binding to the C subunit (32). Because the α4-bound PP2Ac is inactive (7), and α4 cleavage influences PP2Ac levels (Fig. 2), we hypothesize that α4 maintains a reserve pool of inactive PP2Ac at the microtubules that can be quickly incorporated into active PP2A holoenzymes. At the molecular level, increases in intracellular calcium have been shown to promote Tau hyperphosphorylation (33). Given that α4 cleavage is mediated by calpains, which can be hyperactivated in response to elevated calcium flux (34), it is enticing to speculate that the hyperphosphorylation of Tau could result from up-regulated α4 cleavage and a loss of PP2Ac at microtubules. Pharmacological agents that increase microtubule-associated PP2A levels by interfering with α4 monoubiquitination or cleavage are an attractive avenue for the treatment of AD and other tauopathies. Two drugs that should be considered in this regard are metformin and sodium selenate as they have been shown to stabilize Tau-associated PP2A and reduce Tau phosphorylation in cellular and animal models of AD (35–37). In summary, our studies have uncovered a novel regulatory process for PP2A involving ubiquitination-induced cleavage of α4, which plays a crucial role in modulating PP2Ac levels in both normal and pathophysiological conditions.
Supplementary Material
Acknowledgments
We thank Ana Carneiro for expert advice and Elaine Loomis for outstanding technical assistance. We also thank the University of Washington Alzheimer's Disease Research Center (Murray Raskind, Director) for providing tissue samples and the Proteomics Core of the Mass Spectrometry Research Center at Vanderbilt University (David Friedman, Associate Director) for proteomic analyses.
This work was supported by National Institutes of Health Grants DK070787 and GM051366 (to B. E. W.), NS064131 (to B. C. K.), and T32 GM07628 (to G. R. W.); American Cancer Society IRG-58-009-49 (to B. W. S.); Department of Veterans Affairs Merit Review Grant 114891 (to B. C. K.); and Ataxia UK and Tenovus Scotland (to S. S.).

This article contains supplemental “Materials and Methods” and Figs. S1–S3.
- PP2A
- protein phosphatase 2A
- MAP
- microtubule-associated protein
- AD
- Alzheimer disease
- OS
- Opitz syndrome
- PP2Ac
- PP2A catalytic subunit
- HEF
- human embryonic fibroblast
- UIM
- ubiquitin-interacting motif.
REFERENCES
- 1. Trockenbacher A., Suckow V., Foerster J., Winter J., Krauss S., Ropers H. H., Schneider R., Schweiger S. (2001) MID1, mutated in Opitz syndrome, encodes an ubiquitin ligase that targets phosphatase 2A for degradation. Nat. Genet. 29, 287–294 [DOI] [PubMed] [Google Scholar]
- 2. Liu R., Wang J. Z. (2009) Protein phosphatase 2A in Alzheimer disease. Pathophysiology 16, 273–277 [DOI] [PubMed] [Google Scholar]
- 3. Sontag J. M., Sontag E. (2006) Regulation of cell adhesion by PP2A and SV40 small tumor antigen: An important link to cell transformation. Cell Mol. Life Sci. 63, 2979–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Janssens V., Goris J. (2001) Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signaling. Biochem. J. 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Virshup D. M., Shenolikar S. (2009) From promiscuity to precision: Protein phosphatases get a makeover. Mol. Cell 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 6. Liu J., Prickett T. D., Elliott E., Meroni G., Brautigan D. L. (2001) Phosphorylation and microtubule association of the Opitz syndrome protein mid-1 is regulated by protein phosphatase 2A via binding to the regulatory subunit α4. Proc. Natl. Acad. Sci. U.S.A. 98, 6650–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kong M., Ditsworth D., Lindsten T., Thompson C. B. (2009) α4 is an essential regulator of PP2A phosphatase activity. Mol. Cell 36, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. LeNoue-Newton M., Watkins G. R., Zou P., Germane K. L., McCorvey L. R., Wadzinski B. E., Spiller B. W. (2011) The E3 ubiquitin ligase- and protein phosphatase 2A (PP2A)-binding domains of the α4 protein are both required for α4 to inhibit PP2A degradation. J. Biol. Chem. 286, 17665–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McConnell J. L., Watkins G. R., Soss S. E., Franz H. S., McCorvey L. R., Spiller B. W., Chazin W. J., Wadzinski B. E. (2010) α4 is a ubiquitin-binding protein that regulates protein serine/threonine phosphatase 2A ubiquitination. Biochemistry 49, 1713–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quaderi N. A., Schweiger S., Gaudenz K., Franco B., Rugarli E. I., Berger W., Feldman G. J., Volta M., Andolfi G., Gilgenkrantz S., Marion R. W., Hennekam R. C., Opitz J. M., Muenke M., Ropers H. H., Ballabio A. (1997) Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat. Genet. 17, 285–291 [DOI] [PubMed] [Google Scholar]
- 11. Guthrie C. R., Kraemer B. C. (2011) Proteasome inhibition drives HDAC6-dependent recruitment of Tau to aggresomes. J. Mol. Neurosci. 45, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Han X., Du H., Massiah M. A. (2011) Detection and characterization of the in vitro e3 ligase activity of the human MID1 protein. J. Mol. Biol. 407, 505–520 [DOI] [PubMed] [Google Scholar]
- 13. Soss S. E., Yue Y., Dhe-Paganon S., Chazin W. J. (2011) E2 conjugating enzyme selectivity and requirements for function of the E3 ubiquitin ligase CHIP. J. Biol. Chem. 286, 21277–21286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Windheim M., Peggie M., Cohen P. (2008) Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem. J. 409, 723–729 [DOI] [PubMed] [Google Scholar]
- 15. Smetana J. H., Oliveira C. L., Jablonka W., Aguiar Pertinhez T., Carneiro F. R., Montero-Lomeli M., Torriani I., Zanchin N. I. (2006) Low resolution structure of the human α4 protein (IgBP1) and studies on the stability of α4 and of its yeast ortholog Tap42. Biochim Biophys. Acta 1764, 724–734 [DOI] [PubMed] [Google Scholar]
- 16. Yang J., Roe S. M., Prickett T. D., Brautigan D. L., Barford D. (2007) The structure of Tap42/α4 reveals a tetratricopeptide repeat-like fold and provides insights into PP2A regulation. Biochemistry 46, 8807–8815 [DOI] [PubMed] [Google Scholar]
- 17. Yang Y., Kitagaki J., Dai R. M., Tsai Y. C., Lorick K. L., Ludwig R. L., Pierre S. A., Jensen J. P., Davydov I. V., Oberoi P., Li C. C., Kenten J. H., Beutler J. A., Vousden K. H., Weissman A. M. (2007) Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 67, 9472–9481 [DOI] [PubMed] [Google Scholar]
- 18. Hicke L., Schubert H. L., Hill C. P. (2005) Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 19. Kong M., Fox C. J., Mu J., Solt L., Xu A., Cinalli R. M., Birnbaum M. J., Lindsten T., Thompson C. B. (2004) The PP2A-associated protein α4 is an essential inhibitor of apoptosis. Science 306, 695–698 [DOI] [PubMed] [Google Scholar]
- 20. Schweiger S., Foerster J., Lehmann T., Suckow V., Muller Y. A., Walter G., Davies T., Porter H., van Bokhoven H., Lunt P. W., Traub P., Ropers H. H. (1999) The Opitz syndrome gene product, MID1, associates with microtubules. Proc. Natl. Acad. Sci. U.S.A. 96, 2794–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aranda-Orgillés B., Aigner J., Kunath M., Lurz R., Schneider R., Schweiger S. (2008) Active transport of the ubiquitin ligase MID1 along the microtubules is regulated by protein phosphatase 2A. PLoS One 3, e3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goedert M., Jakes R., Qi Z., Wang J. H., Cohen P. (1995) Protein phosphatase 2A is the major enzyme in brain that dephosphorylates Tau protein phosphorylated by proline-directed protein kinases or cyclic AMP-dependent protein kinase. J. Neurochem. 65, 2804–2807 [DOI] [PubMed] [Google Scholar]
- 23. Selkoe D. J. (2001) Alzheimer disease: Genes, proteins, and therapy. Physiol. Rev. 81, 741–766 [DOI] [PubMed] [Google Scholar]
- 24. Ittner L. M., Götz J. (2011) Amyloid-β and Tau–a toxic pas de deux in Alzheimer disease. Nat. Rev. Neurosci. 12, 65–72 [DOI] [PubMed] [Google Scholar]
- 25. Haass C., Mandelkow E. (2010) Fyn-Tau-amyloid: A toxic triad. Cell 142, 356–358 [DOI] [PubMed] [Google Scholar]
- 26. Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 27. Twomey C., Qian S., McCarthy J. V. (2009) TRAF6 promotes ubiquitination and regulated intramembrane proteolysis of IL-1R1. Biochem. Biophys. Res. Commun. 381, 418–423 [DOI] [PubMed] [Google Scholar]
- 28. Gupta-Rossi N., Six E., LeBail O., Logeat F., Chastagner P., Olry A., Israel A., Brou C. (2004) Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J. Cell Biol. 166, 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stabach P. R., Cianci C. D., Glantz S. B., Zhang Z., Morrow J. S. (1997) Site-directed mutagenesis of αII spectrin at codon 1175 modulates its mu-calpain susceptibility. Biochemistry 36, 57–65 [DOI] [PubMed] [Google Scholar]
- 30. Xu Y., Chen Y., Zhang P., Jeffrey P. D., Shi Y. (2008) Structure of a protein phosphatase 2A holoenzyme: Insights into B55-mediated Tau dephosphorylation. Mol. Cell 31, 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sontag E., Nunbhakdi-Craig V., Lee G., Bloom G. S., Mumby M. C. (1996) Regulation of the phosphorylation state and microtubule-binding activity of Tau by protein phosphatase 2A. Neuron. 17, 1201–1207 [DOI] [PubMed] [Google Scholar]
- 32. Prickett T. D., Brautigan D. L. (2004) Overlapping binding sites in protein phosphatase 2A for association with regulatory A and α-4 (mTap42) subunits. J. Biol. Chem. 279, 38912–38920 [DOI] [PubMed] [Google Scholar]
- 33. Zempel H., Thies E., Mandelkow E., Mandelkow E. M. (2010) Abeta oligomers cause localized Ca2+ elevation, missorting of endogenous Tau into dendrites, Tau phosphorylation, and destruction of microtubules and spines. J. Neurosci. 30, 11938–11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kambe A., Yokota M., Saido T. C., Satokata I., Fujikawa H., Tabuchi S., Kamitani H., Watanabe T. (2005) Spatial resolution of calpain-catalyzed proteolysis in focal cerebral ischemia. Brain Res. 1040, 36–43 [DOI] [PubMed] [Google Scholar]
- 35. Kickstein E., Krauss S., Thornhill P., Rutschow D., Zeller R., Sharkey J., Williamson R., Fuchs M., Köhler A., Glossmann H., Schneider R., Sutherland C., Schweiger S. (2010) Biguanide metformin acts on Tau phosphorylation via mTOR/protein phosphatase 2A (PP2A) signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 21830–21835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Corcoran N. M., Martin D., Hutter-Paier B., Windisch M., Nguyen T., Nheu L., Sundstrom L. E., Costello A. J., Hovens C. M. (2010) Sodium selenate specifically activates PP2A phosphatase, dephosphorylates Tau and reverses memory deficits in an Alzheimer disease model. J. Clin. Neurosci. 17, 1025–1033 [DOI] [PubMed] [Google Scholar]
- 37. van Eersel J., Ke Y. D., Liu X., Delerue F., Kril J. J., Götz J., Ittner L. M. (2010) Sodium selenate mitigates Tau pathology, neurodegeneration, and functional deficits in Alzheimer disease models. Proc. Natl. Acad. Sci. U.S.A. 107, 13888–13893 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.