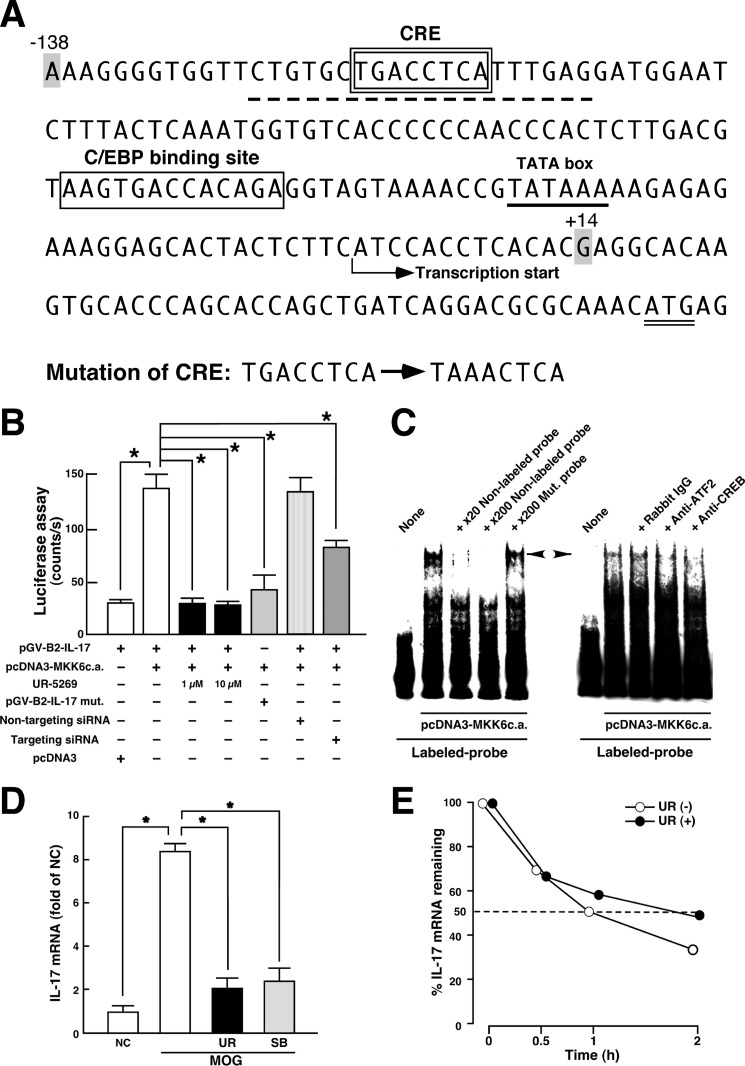
FIGURE 4.
Transcriptional regulation of IL-17 production by p38 pathway. A, nucleotide sequence of 5′-flanking proximal region of mouse Il17a gene. CRE- and C/EBP-binding site are enclosed by a double-line and a single-line square, respectively. The TATA box (underline), initiation codon (double underline), probe sequence used in C (dotted underline), and transcriptional start site are indicated. The mutation sites produced in CRE are described. The region (−138/+14) of mouse Il17a gene was subcloned into pGV-B2 luciferase reporter vector. B, luciferase assay for p38 signal-dependent regulation of Il17a gene. Jurkat cells were transfected with the indicated expression plasmids. siRNAs were introduced into the cells 48 h before the transfection of plasmids. Cell lysates were subjected to luciferase assay 24 h after incubation. To determine the effect of UR-5269, incubation was performed in the presence of 1 or 10 μm UR-5269. Data are shown as mean ± S.E. (n = 6). *, p < 0.05, significantly different from stimulation value determined as co-transfection of pGV-B2-IL-17 and pcDNA3-MKK6c.a. (ANOVA with Bonferroni method). C, EMSA for p38-induced protein-probe complexes. Nuclear extracts were prepared from Jurkat cells transfected with pcDNA3-MKK6c.a. and subjected to interaction with labeled WT probe (dotted underline in A). Competition assays were performed by further adding 20- and 200-fold molar excess of unlabeled WT probe or 200-fold molar excess of unlabeled Mut. probe (mutation sites in the CRE as described in A). A left-directed arrowhead indicates specific band showing the protein complexes bound to the CRE in proximal promoter of Il17 gene. A right-directed arrowhead indicates a disappeared band in EMSA when nuclear extracts were preincubated with either anti-ATF2 antibody or anti-CREB antibody. Similar results were obtained from three independent experiments. D, effect of p38 inhibitors on MOG(35–55)-induced IL-17 mRNA expression in LNC. LNC from WT mice at 8 dpi were stimulated in vitro with MOG(35–55) for 24 h in the absence or presence of p38 inhibitor (UR-5269 (UR), black bars; SB203580 (SB), shaded bars). Total RNA was subjected to a real time RT-PCR for IL-17. Signal values were normalized by the expression of β-actin and expressed as fold induction over the negative control (NC). Data are shown as mean ± S.E. (n = 6). *, p < 0.05, significantly different from stimulation value (ANOVA with Bonferroni method). E, effect of p38 on IL-17 mRNA stability. Th17 cells differentiated in vitro were stimulated with phorbol 12-myristate 13-acetate and ionomycin in the presence (closed circle) or absence (open circle) of 1 μm UR-5269 for 30 min, and then actinomycin D (5 μg/ml) was added for 0.5, 1, and 2 h. Total RNA was subjected to a real time RT-PCR for IL-17. Results are expressed as percentage of IL-17 mRNA remaining after normalized to β-actin. Means from two independent experiments are presented.