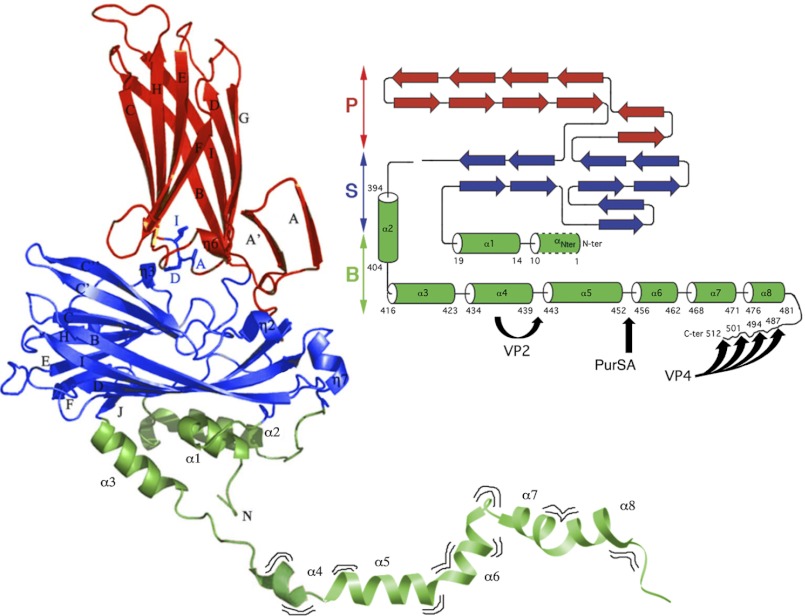
FIGURE 1.
Structure and maturation of pVP2, the capsid protein precursor of IBDV. X-ray structure of VP2 (PDB code 2GSY) manually bound (at residue 441) to part of the pVP2 C terminus determined by NMR analysis (segment 442–488, PDB code 2IMU). Domains S, P and B are colored red, blue, and green, respectively. The VP2 atomic structure extends up to residue 441, including the highly flexible helix α4. The amphipathic α-helix (residues 443–452) is marked as helix α5. The α8 helix ends at residue 481, and the most C-terminal residues assumed display a disordered conformation. Wavy lines indicate that the pVP2 C-terminal extension exhibits conformational flexibility. On the right, a simplified topology schematic (cylinders, α-helices; arrows, β-strands) are shown color-coded by domains. Black arrows indicate processing sites of protease activities performed by VP4, VP2, and PurSA, respectively. VP4 targets: Ala487-Ala488, Ala494-Ala495, Ala501-Ala502 and Ala512-Ala513. VP2 targets: Ala441-Phe442. PurSA targets (this study): Arg452-Arg453.