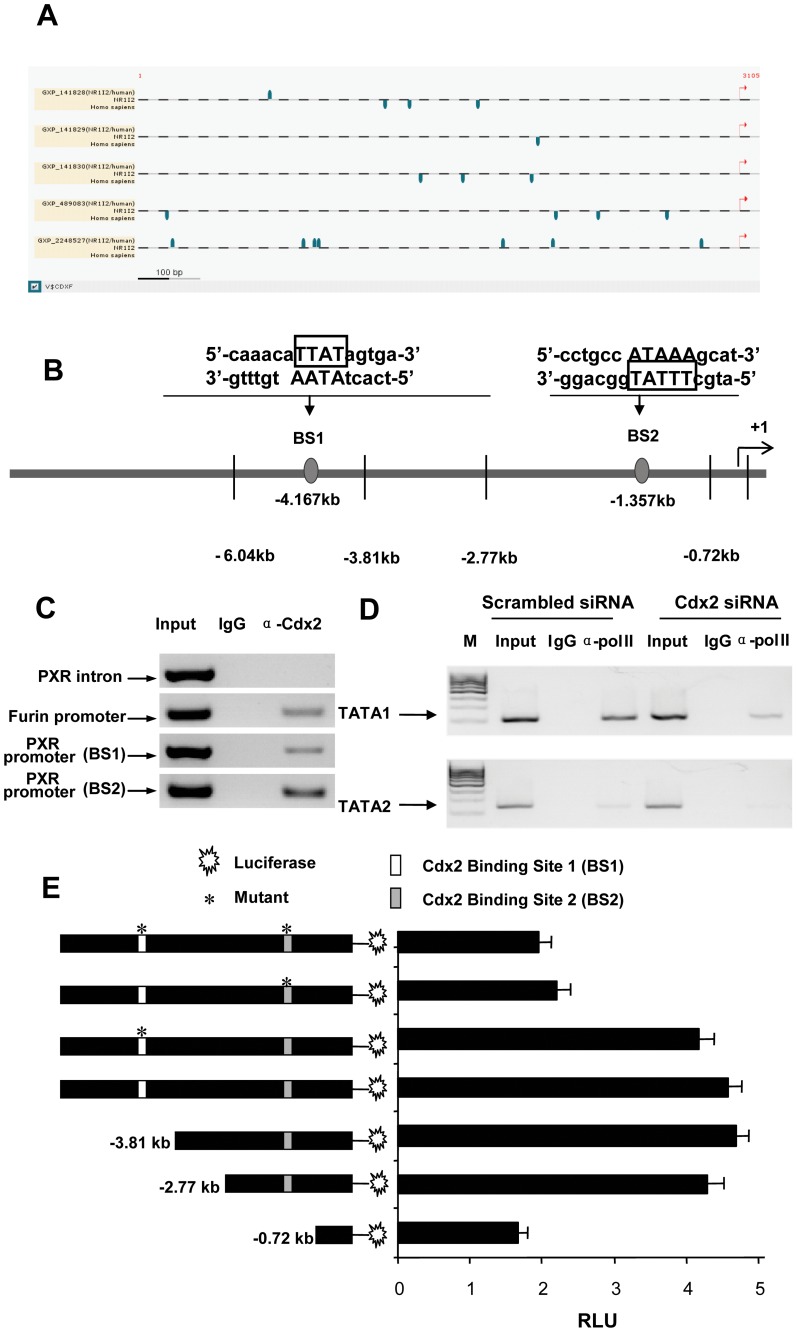
Figure 6. Cdx2 binds to two specific endogenous binding sites on the proximal PXR promoter.
(A) Schematic details of the sites (blue marker) that were identified with a high match [matrix scores (≥0.85)], represent potential functional Cdx2 binding sites on PXR promoter. (Note: three potential Cdx1 binding sites, which are also identified on analysis, were excluded from consideration). The in silico analysis identified a total of 19 potential Cdx1/2 binding sites, of which, we verified endogenous binding for two sites, BS1 and BS2 by (C) Chromatin Immunoprecipitation (ChIP) assays. BS, Cdx2 binding site. ChIP assays were performed with Cdx2 and non-specific IgG antibodies and Cdx2 binding was assessed on PXR promoter (BS1 and BS2), PXR intron (negative control) and Furin promoter (positive control) regions. (D) Pol II ChIP assays were performed, using chromatin extracted from LS174T cells that were exposed to scrambled or Cdx2 siRNA (si-Cdx2). Pol II occupancy was assessed on PXR proximal promoter (TATA box) region, using Pol II and non-specific IgG antibodies as a control. (E) Cdx2 transactivation assay was performed in 293T cells, transfected with ∼6 kb PXR promoter reporter constructs (with or without deletion and mutations, as shown in the figure) and Cdx2 expression plasmid. Data expressed as RLU. RLU, relative light unit. Histogram, mean ± SD.