Figure 1.
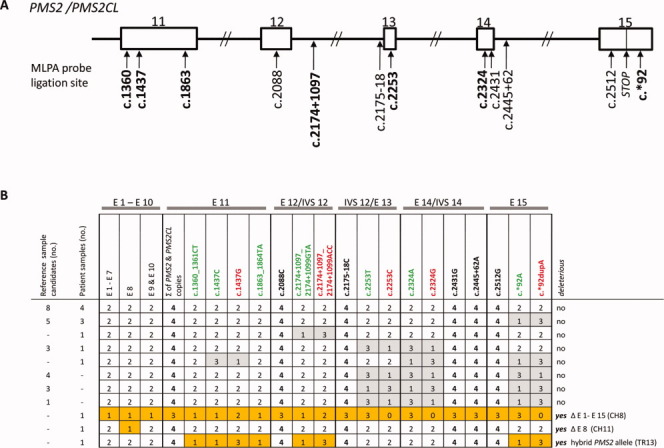
PMS2 MLPA results for reference DNA candidates and DNA samples from patients with colorectal tumors displaying isolated PMS2 expression loss. (A) Schematic showing ligation sites for the probes in the redesigned (P008-B1) PMS2 MLPA kit. Paralog-discriminating ligation sites are shown in boldface. Rectangles: exons 11–15 of PMS2 (and PMS2CL); lines: introns. (B) MLPA results (given as copy numbers of the DNA sequences bound by the MLPA probes) for 24 reference DNA sample candidates (obtained from Coriell Cell Repositories) and 13 patient DNA samples. Results are grouped for exons 1–7 (E1-E7) and 9 and 10 (E9 and E10) and reported individually for exons 8 (E8) and exons 11 (E11) to 15 (E15), the latter being shared by the paralogs. The paralog-discriminating sequence variants at the ligation sites of the probes in exons 11–15 are color-coded: green, PMS2-specific probes; red, PMS2CL-specific probes; black, nonspecific probes. Numbers in cells (0–4): copies of exon sequences in sample. Paired gray cells: uneven distribution of PMS2- and PMS2CL-specific signals in samples with a total of four copies, which reflects the presence of nondeleterious hybrid alleles (PMS2 or PMS2CL). Orange cells: aberrant MLPA results in patient CH8 (reflecting deletion of all PMS2 exons); patient CH11 (exon 8 deletion); and patient TR13 (who had a deleterious PMS2 hybrid allele; see text for details). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
