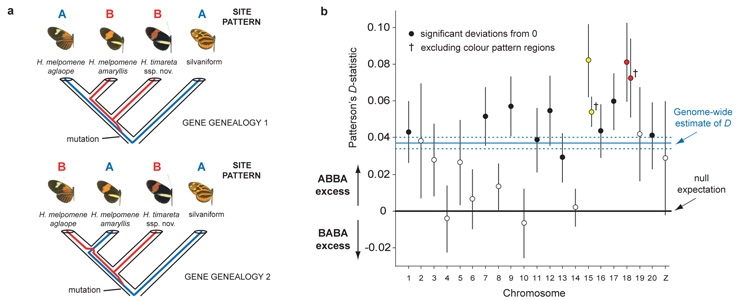
Figure 3. Four-taxon ABBA-BABA test of introgression.
a ABBA and BABA nucleotide sites employed in the test are derived (– – B –) in H. timareta, compared with the silvaniform outgroup (– – – A), but differ among H. melpomene amaryllis and H. melpomene aglaope (either ABBA or BABA). As this almost exclusively restricts attention to sites polymorphic in the ancestor of H. timareta and H. melpomene, equal numbers of ABBA and BABA sites22 are expected under a null hypothesis of no introgression, as depicted in the two gene genealogies. b Distribution among chromosomes of Patterson’s D-statistic ± s.e., which measures excess of ABBA vs. BABA sites22, here for the comparison H. m. aglaope; H. m. amaryllis; H. timareta ssp. nov.; silvaniform. Chromosomes containing the two colour pattern regions (B/D red; N/Yb yellow) have the two highest D-statistics; the combinatorial probability of this occurring by chance is 0.005. The excess of ABBA sites (0 < D < 1) indicates introgression between sympatric H. timareta and H. melpomene amaryllis.