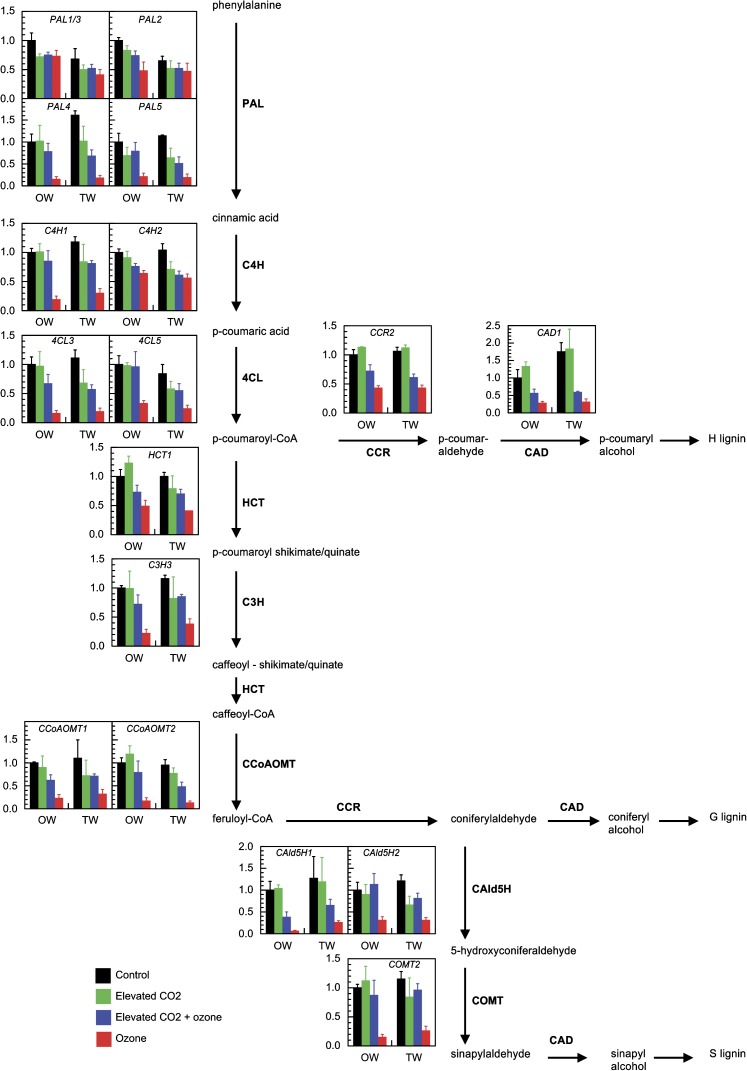
Fig. 3.
Transcript abundance of genes involved in the lignin biosynthesis pathway. Analyses were performed on opposite wood (OW) and tension wood (TW) of lower stem of hybrid poplar subjected for 60 d to different treatments. Transcript levels were normalized and expressed as fold changes compared with values of opposite wood in control conditions. y axis is in arbitrary units. Values are mean ±SE (n=3 biological replicates). For statistical differences refer to Supplementary Table S2 at JXB online. The most favoured route in angiosperms lignin pathway is shown. Phenylalanine ammonia-lyase (PAL), cinnamate-4-hydroxylase (C4H), 4-coumarate:CoA ligase (4CL), p-hydroxycinnamoyl-CoA:quinate/shikimate p-hydroxycinnamoyltransferase (HCT), 4-coumarate 3-hydroxylase (C3H), caffeoyl-CoA O-methyltransferase (CCoAOMT), cinnamoyl-CoA reductase (CCR), coniferyl aldehyde 5-hydroxylase (CAld5H), caffeic acid/5-hydroxyconiferaldehyde O-methyltransferase (COMT), cinnamyl alcohol dehydrogenase (CAD).