Figure 3.
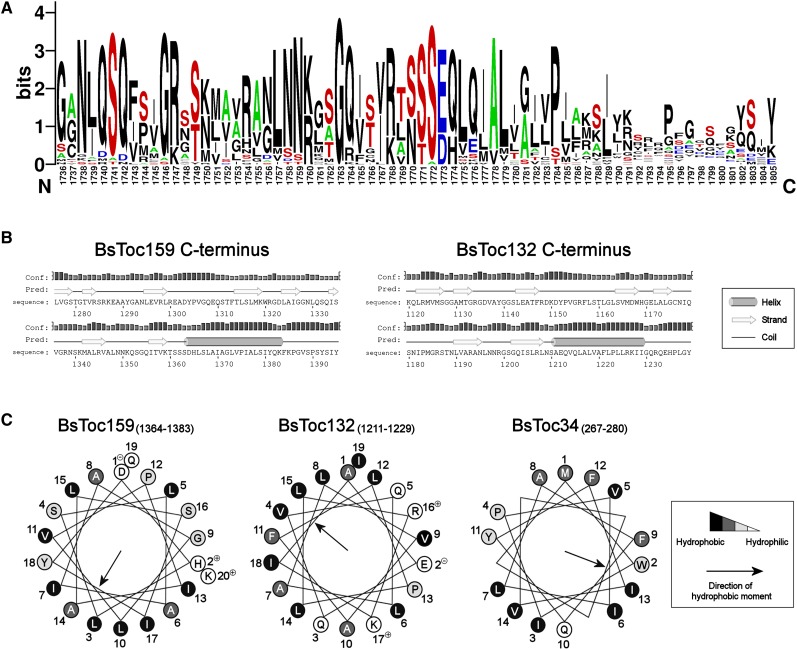
Bioinformatics Prediction of a cTP-Like Targeting Signal at the C terminus of Toc159.
(A) WebLogo representation of the aligned C-terminal sequences from Toc159 homologs. Amino acid sequence alignment was performed using the ClustalW algorithm and is available as Supplemental Data Set 1 online. Sequence logos were generated from the alignment using WebLogo 2.8.2 (Crooks et al., 2004). Letter height is proportional to its frequency of occurrence. The graph illustrates high frequencies of hydroxylated residues (in red) and Ala residues (in green) and low frequencies of acidic residues (in blue).
(B) Prediction of secondary structures at the C termini of Toc159 homologs. Predictions were performed using the Web-based program PSIPRED v3.0 (Jones, 1999). The height of the bar for each residue is proportional to the confidence level of prediction.
(C) Helical wheel projections of the predicted C-terminal helices of Toc159 homologs and the transmembrane helix of Toc34. Numbering designates the order of amino acid sequence. Hydrophobicities of residues are highlighted on the Kyte-Doolittle hydropathy scale (hydrophilic in white, neutral in light gray, hydrophobic in dark gray, and very hydrophobic in black).
[See online article for color version of this figure.]