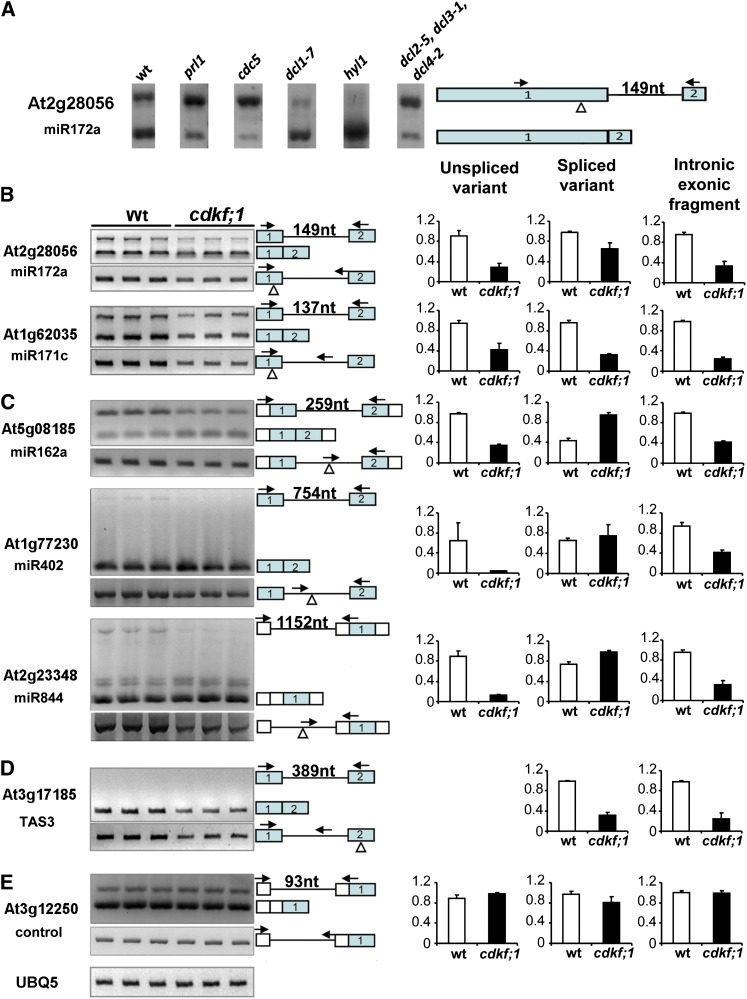
Figure 8.
The cdkf;1 Mutation Enhances the Splicing of Pre-miRNA–Containing Introns.
(A) Comparison of intron splicing efficiency of pri-miRNA172a in wild-type (wt), prl1, cdc5, dcl1-7, hyl1, and dcl2-5 dcl3-1 dcl4-2 seedlings by RT-PCR. nt, nucleotides.
(B) to (D) RT-PCR assay (left) and qRT-PCR measurements (right).
(B) The levels of introns retained in the vicinity of exonic stem-loops of pre-mRNAs 172a and 171c compared with the level of spliced pri-mRNA isoforms.
(C) Intronic pre-miRNAs 162a, 402, and 844 compared with spliced versions of corresponding transcripts.
(D) Spliced and unspliced forms of TAS3 ta-siRNA precursor.
(E) Normalization of RT- and qRT-PCR assays with the intronless UBI5 (At3g12250) RNA control. Triangles indicate the positions of pre-mRNA stem-loops, arrows mark the positions of exonic and intronic gene-specific primers, and the lengths of introns in nucleotides are indicated. The error bars represent sd of the mean of triplicate qRT-PCR measurements using three biological replicates.
[See online article for color version of this figure.]