Abstract
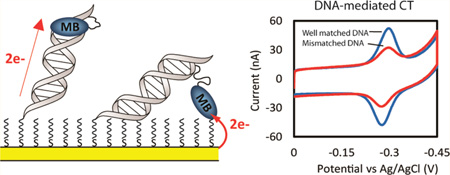
Methylene blue (MB′), covalently attached to DNA through a flexible C12 alkyl linker, provides a sensitive redox reporter in DNA electrochemistry measurements. Tethered, intercalated MB′ is reduced through DNA-mediated charge transport; the incorporation of a single base mismatch at position 3, 10, or 14 of a 17-mer causes an attenuation of the signal to 62 ± 3% of the well-matched DNA, irrespective of position in the duplex. The redox signal intensity for MB′–DNA is found to be least 3-fold larger than that of Nile blue (NB)–DNA, indicating that MB′ is even more strongly coupled to the π-stack. The signal attenuation due to an intervening mismatch does, however, depend on DNA film density and the backfilling agent used to passivate the surface. These results highlight two mechanisms for reduction of MB′ on the DNA-modified electrode: reduction mediated by the DNA base pair stack and direct surface reduction of MB′ at the electrode. These two mechanisms are distinguished by their rates of electron transfer that differ by 20-fold. The extent of direct reduction at the surface can be controlled by assembly and buffer conditions.
INTRODUCTION
Since the discovery that DNA can efficiently serve to conduct electrical current, its properties have been exploited across numerous platforms.1–14 DNA-mediated charge transport (CT) is exquisitely sensitive to perturbations in the intervening base stack, including single base mismatches, lesions, or structural changes caused by proteins.14–16
Electrochemistry experiments on DNA-modified electrodes have been particularly valuable in probing ground state DNA CT and in the development of new DNA-based sensors.11–19 To study and exploit the sensitivity of DNA CT, it is essential that the redox moiety makes an electronic interaction with the base stack of the DNA.17,20
Over the past decade, various reporters have been used for DNA electrochemistry; some redox reporters have been well coupled and others not at all.14–38 Early applications of DNA-modified electrodes primarily depended on the use of noncovalent redox reporters. Noncovalent methylene blue (MB) was widely used as a reporter for DNA CT as it intercalates into the base stack.11,13 Studies with noncovalent MB demonstrated the exquisite sensitivity of the π-stack to subtle perturbations, and MB had the additional advantage that it undergoes electrocatalytic signal amplification.38
However, noncovalent reporters incurred a number of constraints, including the inability to control probe placement and the stringent requirement for high quality DNA films. As such, covalent tethering has been explored to address these issues. Covalent tethering of the reporter results in its restricted mobility; therefore, it must be ensured that coupling between the reporter and base stack is still feasible. The degree of coupling varies based on the mechanism of interaction between the reporter and the base stack.17 Coupling to the base stack has been shown to be possible through various mechanisms including intercalation,18 end-capping,20 and direct conjugation to a nucleic acid base.17 The degree to which the reporter is coupled to the base stack determines the overall efficiency of DNA CT.
Daunomycin, when covalently tethered to the DNA through a short alkyl linkage, has been shown to yield exceptionally strong electrochemical signals.14,16,21 This evidence of strong coupling agrees with the crystal structure that shows daunomycin is tightly intercalated into the base stack.39,40 Despite the exceptional coupling of daunomycin, the field of DNA-mediated CT sensors has evolved to use other reporters as DNA modified with daunomycin is difficult to prepare, is unstable, and has sequence constraints.
More recently, Nile blue (NB) has been used as an electrochemical reporter of DNA CT.12,17 The preparation of NB–DNA does not possess the synthetic limitations of daunomycin, and it is well coupled to the base stack through direct conjugation of NB with a modified uracil.12 In the present study, we built upon this work by developing a system to covalently tether MB, as it has previously been shown to strongly intercalate into the base stack.
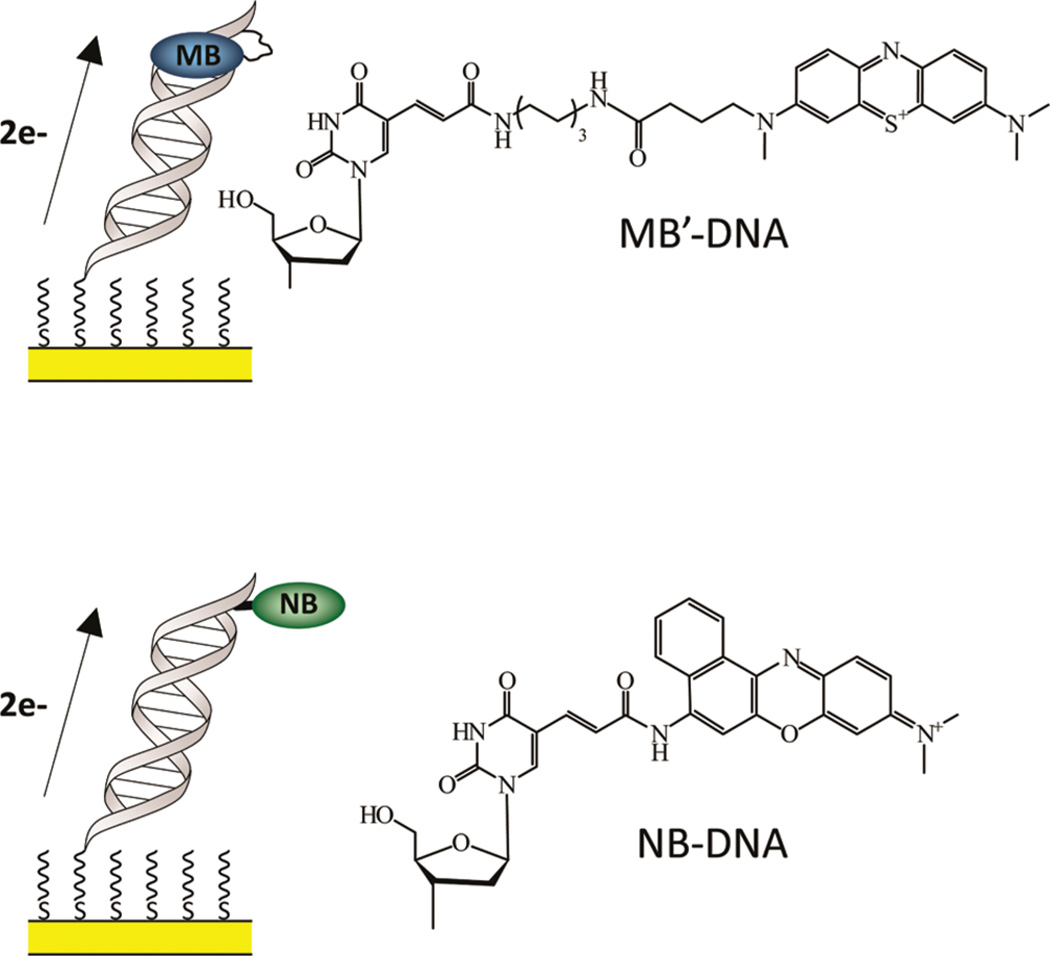
MB′–DNA was tethered through a flexible C12 alkyl linkage to a modified uracil (Figure 1). This linkage further improves the coupling of the reporter by providing the conformational freedom for MB′ to interact with the base stack through intercalation. However, with the enhanced flexibility of the reporter, its ability to be reduced directly at the surface of the electrode must be taken into account.
Figure 1.
Left: schematic illustration of MB′–DNA (top) and NB–DNA (bottom) monolayers bound to an electrode. The reporter is appended to the distal base of the duplex. The intended path for electrochemical reduction is indicated. Right: the chemical structure of both MB′ and NB covalently tethered to a modified uracil.
Various probes have been covalently tethered to the DNA through flexible alkyl linkages and shown to be reduced directly at the electrode surface, including both MB and ferrocene.20,22–37 This has led to a new class of DNA-modified biosensors based upon binding of either oligonucleotides22–32 or DNA-binding proteins33 that cause conformational changes that attenuate the observed signal by physically diminishing the surface accessibility of the reporter. These biosensors are based on the surface reduction of the reporter, and thus the reporter does not couple with the base stack. Ferrocene has been shown to be a common reporter for this platform given its poor coupling to the base stack and yielding efficient reduction at the electrode surface.37 More recently, MB-modified DNA has been frequently used.22–33
In this work, the mechanism of reduction for MB′ covalently tethered to DNA through a flexible C12 alkyl linkage to a modified uracil is investigated (Figure 1). MB′–DNA, covalently tethered through this flexible linkage, couples to the base stack through intercalation. This is compared to the previously established reporter, NB–DNA, which is covalently tethered through a short rigid linkage and couples to the base stack through direct conjugation. MB′–DNA is shown not only to be reduced via DNA CT but is also capable of being reduced directly at the surface. We describe the conditions under which DNA CT is the primary mechanism for MB′ reduction.
EXPERIMENTAL SECTION
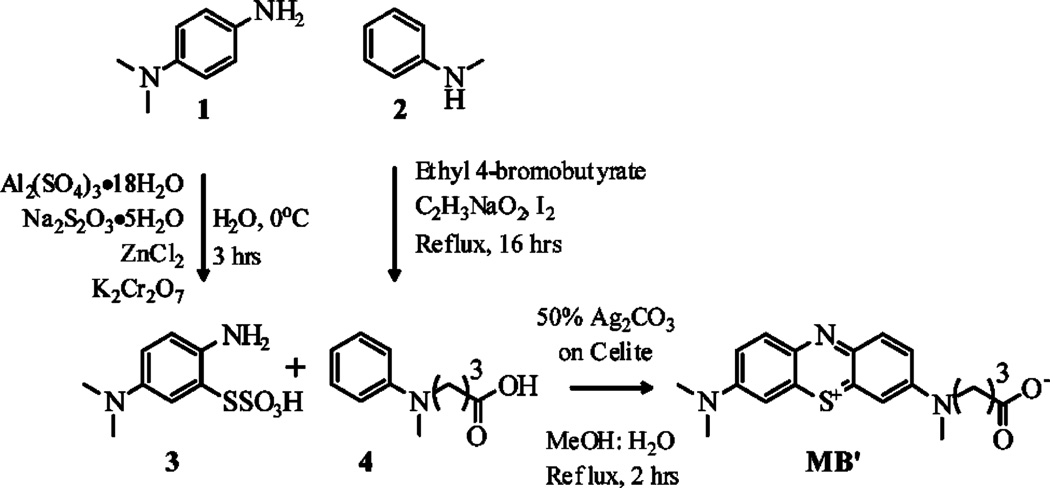
Synthesis of Modified NHS Ester Activated Methylene Blue′ (Scheme 1)
Scheme 1.
Synthetic Strategy for the Preparation of N-Carboxypropyl Methylene Blue (MB′)
All materials were used as purchased from Sigma-Aldrich.
2-Amino-5-(dimethylamino)phenylthiosulfonic Acid (3) Preparation
3 was prepared according to the procedure described by Wanger41 by separately dissolving aluminum sulfate octadecahydrate (43.6 mg, 65 mmol), sodium thiosulfate (22.0 g, 140 mmol), and zinc chloride (8.8 g, 63 mmol) in 100, 80, and 12 mL of water, respectively, and added to N,N-dimethylphenylenediamine (1) (10 g, 73 mmol) in a 500 mL round-bottom flask. The reaction mixture was then cooled to 0 °C while continuously stirring. Potassium dichromate (5.0 g, 17 mmol) was dissolved in 30 mL of water and added dropwise to the reaction mixture over 15 min. The reaction was kept at 0 °C for 2 h and then slowly allowed to warm to room temperature. The precipitate was isolated by vacuum distillation and washed with water, acetone, and ether. The purple solid (7.4 g, 30 mmol, 41% yield) was confirmed as the desired product by 1H NMR (DMSO-d6) and was used in subsequent reactions without further purification.
N-Methyl-N-(carboxypropyl)aniline (4) Preparation
4 was prepared according to the procedure described by Whitten.42 N-Methylaniline (2) (15.17 mL, 140 mmol) was refluxed with ethyl-4-bromobutyrate (20.0 mL, 140 mmol) for 16 h in a 100 mL round-bottom flask. The reaction mixture was then cooled to room temperature, and water (15 mL) was added. The crude reaction mixture was then made basic by the dropwise addition of saturated sodium hydroxide and extracted with ether (3 × 50 mL). The extract was washed with water and dried over MgSO4, and the ether was removed under reduced pressure. Pure N-methyl-N-ethyl-4-butanoate aniline (16.3 g, 74 mmol, 53% yield) was isolated, as a clear liquid, by vacuum filtration at 110 °C at 0.2 mmHg. The product was confirmed by 1H NMR and 13C NMR (CDCl3) and an observed mass of 221.3 g/mol (calculated mass of 221.3 g/mol) in ESI-MS in acetonitrile: water:acetic acid (1:1:0.1%).
The ester product (9.4 g, 43 mmol) was subsequently hydrolyzed in 5% KOH (150 mL) by refluxing for 2 h to form the desired carboxylic acid. The reaction was cooled to room temperature and washed with ether (2 × 70 mL). Concentrated hydrochloric acid was added dropwise to the aqueous layer to adjust the pH to 5.5, and then the product was extracted with ether (3 × 100 mL) and dried over MgSO4, and the solvent was removed under reduced pressure. The desired product, 4 (5.1 g, 26 mmol, 62% yield), was confirmed by 1H NMR and 13C NMR (CDCl3) and an observed mass of 193.2 g/mol (calculated mass of 193.2 g/mol) with ESI-MS in acetonitrile:water:-acetic acid (1:1:0.1%).
N-(Carboxypropyl)methlyene Blue (MB′) Preparation
MB′ was prepared by an adapted procedure from Wagner.41 3 (2.4 g, 9.7 mmol) and 4 (1.87 g, 9.7 mmol) were combined and dissolved in a methanol:water mixture (200 mL:80 mL). The reaction was heated to just below reflux (60 °C), and 50% w/w silver carbonate on Celite (10 g) was slowly added. The reaction was then refluxed for 2 h. The reaction was left to cool to room temperature and was vacuum filtered, and the solvent was removed under reduced pressure. The desired product MB′ (0.9 g, 2.5 mmol, 26% yield) was isolated by dry chromatography43 as conventional chromatography techniques were unsuccessful. Impurities were eluted using 20 mL portions of chloroform:methanol:acetic acid (100:15:1.5), and the blue band (MB′) was eluted using portions of chloroform:methanol:acetic acid (100:30:1.5). The final product was confirmed by an observed mass of 356.3 g/mol (calculated mass of 357.13 g/mol) with ESI-MS in acetonitrile:water:acetic acid (1:1:0.1%).
MB′–NHS Ester Preparation
MB′ (8 mg, 0.022 mmol) was dissolved in DMF (1 mL) and combined with N,N′-dicyclohexylcarbodiimide (9.3 mg, 0.045 mmol) and N-hydroxysuccinimide (5.2 mg, 0.045 mmol). The reaction proceeded at room temperature for 24 h. The solvent was removed under reduced pressure, and the material was resuspended in DMSO. Successful ester activation was confirmed by a mass of 454.5 g/mol (calculated mass of 454.54 g/mol) in ESI-MS in acetonitrile:water:acetic acid (1:1:0.1%). Activated ester was not found to be stable for extended periods of time; therefore, it was freshly prepared directly before tethering to amino-modified DNA.
Synthesis of Modified Oligonucleotides
The synthesis and purification of NB and thiol-modified oligonucleotides were carried out following the previously reported protocol.17 Thiol-modified and NHS ester uracil analogue phosphoramidites were purchased from Glen Research.
MB′-modified oligonucleotides were synthesized similarly to NB–DNA with the substitution of an amino-C6-uracil analogue purchased from Glen Research. Amino-modified DNA was purified using standard protocols and then coupled in solution to MB′–NHS ester.
Amino-modified DNA was suspended in 200 µL of a 0.1 M NaHCO3 solution in order to buffer the reaction to a pH of 8.3–8.4. MB′–NHS ester was suspended in DMSO (100 µL) and added to the DNA solution in roughly a 10-fold excess of MB′–NHS ester to amino-modified DNA. The reaction was left to proceed for 12–24 h. A final round of purification was performed by high-performance liquid chromatography (HPLC) using a 50 mM ammonium acetate buffer/acetonitrile gradient with a PLRP-S Column (Agilent). The MB′–DNA mass was confirmed by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. A mass of 5695 g/mol was found for well-matched MB′–DNA, agreeing with with the calculated mass of 5695 g/mol (see Figure 2).
Figure 2.
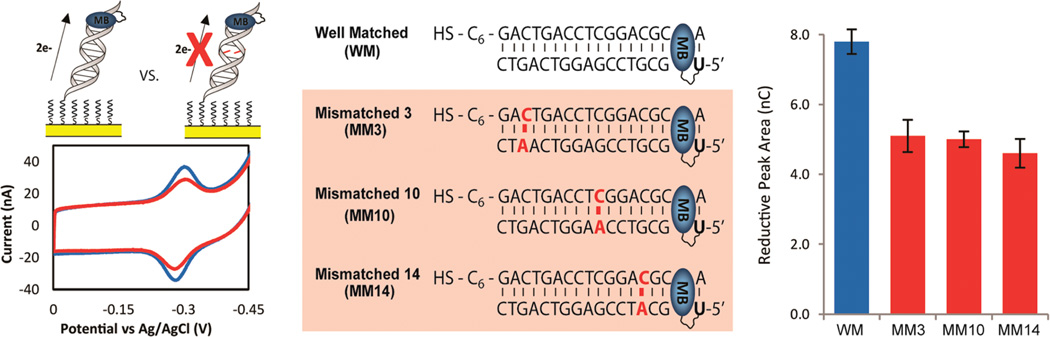
Electrochemistry of MB′–DNA with a single intervening CA mismatch. Left: cyclic voltammetry was acquired at 100 mV/s with either a well-matched sequence (blue) or a sequence containing a single mismatch (red). All four sequences (middle) of MB′–DNA were assembled on the same multiplexed chip and assembled without MgCl2, passivated with MHA, and scanned in spermidine buffer. Right: the area of the reductive signal was used to quantify the signals observed, and the errors denoted are determined from the variation across four different electrodes.
DNA stock solutions were prepared in low salt buffer (5.0 mM phosphate, 50 mM NaCl, pH 7) and quantified as previously reported. The extinction coefficient for single-stranded MB′-DNA at 260 nm was corrected for the absorbance of MB′. This correction was performed by adding the extinction coefficient of MB′ at 260 nm (10 300 M−1 cm−1) to the calculated extinction coefficient for single-stranded DNA. All DNA solutions were thoroughly deoxygenated with argon prior to annealing. Equimolar amounts of thiol-modified and probe-modified oligonucleotides were combined and annealed by heating to 90 °C and cooling to ambient temperature over 90 min to form duplexes.
Preparation of DNA Monolayers and Electrochemical Measurements
Multiplex chips, previously reported,17 were employed for the electrochemical experiments. Each chip contains 16 gold electrodes (2 mm2 area) which were prepared with up to four different kinds of DNA. Equimolar amounts of single-stranded thiol-modified and probe-modified DNA were annealed prior to the electrode assembly. HPLC analysis of duplex DNA stocks were performed prior to electrode assembly to ensure that there were no single-stranded impurities (data not shown). The duplex DNA (25 µL of 25 µM) was then assembled on the electrode surface overnight (20–24 h) in a humid environment to allow for monolayer formation with or without 100 mM MgCl2. Once DNA films were assembled and thoroughly washed with low salt buffer, the electrodes were backfilled with either 1 mM 6-mercaptohexanol (MCH) or 6-mercaptohexanoic acid (MHA) for 45 min in low salt buffer with 5% glycerol. The electrodes were again washed to ensure removal of trace alkanethiols. The electrodes were scanned in a common running buffer of either low salt buffer (5.0 mM phosphate, 50 mM NaCl, and pH 7) or spermidine buffer (5.0 mM phosphate, 50 mM NaCl, 4 mM MgCl2, 4 mM spermidine, 50 µM EDTA, 10% glycerol, and pH 7). An analogous buffer to spermidine buffer was prepared that lacked spermidine, and minimal changes were observed in the electrochemistry (data not shown). Thus, electrochemical differences are best attributed to the effect of adding spermidine. Electrochemical measurements were performed with a CHI620D electrochemical analyzer and a 16-channel multiplexer from CH Instruments. A three-electrode setup was used, with a common Ag/AgCl reference and a Pt wire auxiliary electrodes placed in the central buffer solution.
Cyclic voltammetry (CV) data were collected at 100 mV/s, with the exception of the scan rate dependence experiments where the scan rate is indicated. In order to minimize errors associated with thiol oxidation and surface quality, all CVs overlaid and compared were acquired on the same multiplexed chip with the thiol-modified strand being equivalent in all duplexes.
The film density (Γ) of MB′–DNA and NB–DNA was calculated by the area under the reductive peaks of CVs at 100 mV/s (eq 1).
| (1) |
In eq 1, Q is the area of the reductive signal, n is the number of electrons per redox event (n = 2 for both NB and MB′), F is Faraday’s constant, and A is the area of the gold electrode.
RESULTS
Electrochemistry of MB′–DNA with Intervening Mismatches
In this investigation MB′ has been confined to the distal end of the duplex through a flexible C12-linker appended off the terminally modified uracil (Figure 1). As seen in Figure 2, the resulting CVs from scanning the electrodes in spermidine buffer exhibit strong reductive and oxidative peaks with a midpoint potential of −290 mV versus Ag/AgCl. This is the same reduction potential as freely diffusing MB indicating that covalently tethering MB′ to the DNA through a flexible alkyl chain does not alter its electronic properties.44 The areas of the reductive and oxidative signals were 7.8 ± 0.4 and 7.5 ± 0.3 nC, respectively. Surface-bound species reduced by DNA CT have previously been shown to have cathodic/anodic signals with ratios of nearly unity, which we have ascribed to the fact that the binding affinity of MB for duplex DNA is lowered upon reduction.19,38
In order to demonstrate that the reduction of MB′–DNA occurs via DNA CT, the signal attenuation from the introduction of a single mismatched base pair (CA) intervening between the surface and the probe was compared to that of well matched MB′–DNA. By performing these experiments on multiplexed chips, any variation that can be observed from backfilling the electrodes is removed. Under these conditions, any signal differential obtained between well-matched and mismatched DNA on the same multiplexed chip can be attributed to deficiencies in the CT properties of the mismatched DNA.19 Introduction of a signal base mismatch has been well documented to cause attenuations in DNA CT in both photophysical studies and DNA electrochemistry studies.1–16
Thiol-modified DNA was annealed to the complementary well-matched (WM) DNA sequence and three different mismatched DNA sequences, where the position of the mismatch was varied. The mismatches varied from proximal to the electrode (MM3), in the middle (MM10), and at the distal end (MM14) of the duplex (Figure 2). The four quadrants of a multiplexed chip were used to simultaneously assemble each type of DNA overnight without MgCl2 (Figure 2). In this case the electrodes were passivated by backfilling with MHA and scanned in spermidine buffer. The ratio of the mismatched to well-matched reduction signal areas was quantified in all cases to determine the percent mismatch signal. The signal area of well-matched MB′–DNA was found to be 7.8 ± 0.4 nC. For the various mismatches, the percent signal remaining was 65%, 64%, and 59% for a mismatch incorporated at position 3, 10, or 14, respectively. Under these assembly conditions, the percent signal remaining, due to the incorporation of the CA mismatch, was essentially equivalent regardless of the position in the duplex. The suppression of the MB′ signal from a single CA mismatch validated that MB′ is well coupled to the π-stack and the electrons that reduce MB′ are traveling through the intervening DNA.
It should be noted that we observed signal attenuation from a CA mismatch at the 14th position of the 17-mer, indicating that MB′ must intercalate within 3 base pairs of where it is covalently tethered; if MB′ intercalates further away, there would be no observed signal attenuation. This confinement of MB′ is also consistent with what is predicted from model building. Therefore, when MB′ is tethered to the distal end of the duplex, any perturbation to the base stack occurring below the top 7.2 Å will be reported.
Comparison with NB–DNA
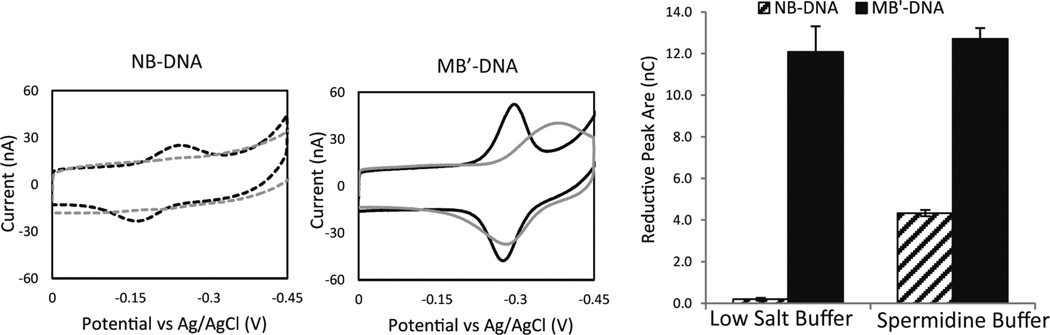
MB′–DNA was then compared with the previously described DNA CT reporter, Nile blue (NB). This comparison used electrodes assembled with 100 mM MgCl2, passivated with MCH, and scanned in spermidine buffer. The area of the reductive signal for MB′–DNA is significantly larger than the signal observed with NB–DNA, 12.5 ± 1.2nC compared to 4.3 ± 0.2 nC, respectively (Figure 3).
Figure 3.
Comparison of the signal from NB–DNA (dotted) and MB′–DNA (solid). The electrodes were assembled with 100 mM MgCl2 and passivated with MCH. CVs were acquired at 100 mV/s, and all electrodes were scanned in both a low salt buffer (5.0 mM phosphate, 50 mM NaCl, and pH 7) (gray) and a spermidine buffer (5.0 mM phosphate, 50 mM NaCl, 4 mM MgCl2, 4 mM spermidine, 50 µM EDTA, 10% glycerol, and pH 7) (black). The CVs shows the overall signal changes in response to the running buffer. The reductive peak areas were quantified to show that the amount of MB′–DNA being reduced is relatively unchanged regardless of the buffer. The errors denoted are determined from the variation across four different electrodes.
Additionally, when the same electrodes are scanned in low salt buffer, NB–DNA has an average signal area of 0.2 ± 0.1 nC (Figure 3), while the reductive peak area for MB′–DNA remains relatively unchanged at 11.8 ± 0.5 nC. Despite the area of MB′ remaining minimally affected, other peak characteristics were significantly altered. Switching to a low salt buffer resulted in a broadening of the peak and an increase in the peak splitting. The sharper peaks in spermidine buffer indicate that the film is more homogeneous. The peak splitting between the oxidative and reductive peaks decreases from 140 mV in low salt buffer to 30 mV in spermidine buffer. These peak changes are largely reversible; there is, however, not a complete restoration of the peak observed in low salt buffer, indicating that the spermidine is not fully removed.
Variation in Running Buffer for Optimized Mismatch Discrimination
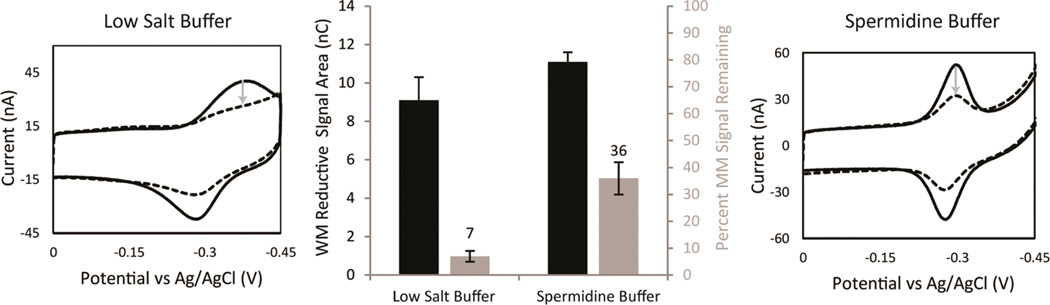
The degree of signal attenuation upon introduction of a single mismatched base (MB′–MM10 DNA) was compared in low salt buffer and spermidine buffer. The DNA was assembled with 100 mM MgCl2 and backfilled with MHA. Well-matched MB′–DNA shows a reductive signal area of 11.1 ± 0.2 nC while mismatched MB′–DNA shows a signal area of 4.0 ± 0.4 nC in spermidine buffer. The percent signal remaining after the introduction of a CA mismatch is 36 ± 6% (Figure 4). When the same experiment is performed with NB–DNA in spermidine buffer, a 50 ± 10% decrease is observed (data not shown). This result agrees with the previously reported values with NB–DNA in that the percent signal remaining due to a CA mismatch ranges from 30 to 60%.12,19
Figure 4.
Optimization of mismatch discrimination depending on the running buffer. Both WM MB′–DNA (solid) and MM10 MB′–DNA (dotted) modified electrodes were assembled with 100 mM MgCl2, passivated with MHA, and scanned in both a low salt buffer (5.0 mM phosphate, 50 mM NaCl, and pH 7) (right) and a spermidine buffer (5.0 mM phosphate, 50 mM NaCl, 4 mM MgCl2, 4 mM spermidine, 50 µM EDTA, 10% glycerol, and pH 7) (left). The areas of the reductive peaks were used to quantify the reductive signal size (black) and determine the percent signal remaining ([MM]/[WM] × 100) (gray) from the incorporation of a single mismatch. The gray arrows denote the decrease in signal area with the optimal percent signal attenuation being in low salt buffer. The errors denoted were determined by the standard deviation from four electrodes averages, for each sequence of DNA, across three chips.
When the same MB′–DNA electrodes were then examined in low salt buffer, the mismatched reductive signal is significantly broadened and decreased in intensity, yielding an area of 0.5 ± 0.1 nC. This broadening decreases the percent signal remaining to only 7 ± 2% of the well-matched signal (9.1 ± 1.2 nC) (Figure 4). This same effect was observed in a buffer equivalent to spermidine buffer but lacking spermidine (data not shown). Spermidine binds in the grooves of duplex DNA, which rigidifies the duplex, resulting in sharper peaks. However, spermidine should also decrease the binding affinity of MB′, which may account for the decreased signal attenuation.
Variation in Assembly Conditions: Density of DNA Film
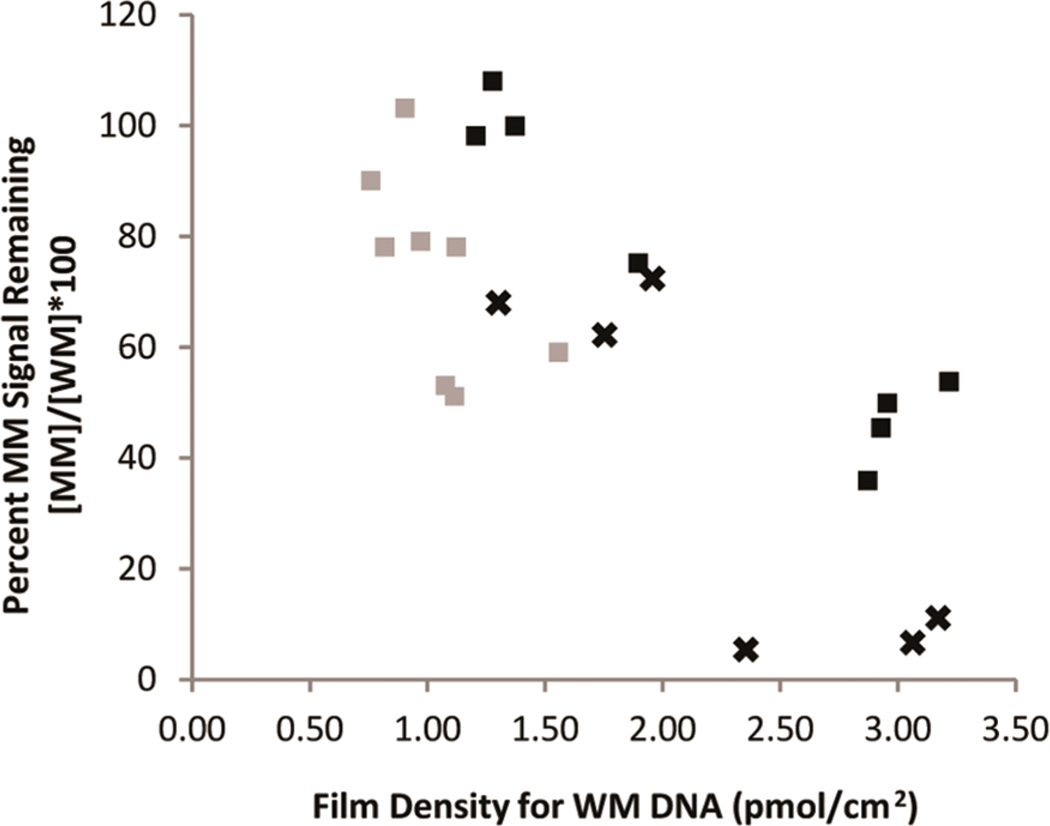
In addition to buffer conditions, the effect of varying the assembly conditions on mismatch discrimination was investigated. The varying densities of DNA were obtained by assembling with and without 100 mM MgCl2. In the absence of MgCl2, the negatively charged phosphate backbone of DNA duplexes repel each other, yielding significantly lower film densities.13,20 MB′–DNA and NB–DNA films were therefore examined with and without incubation with 100 mM MgCl2. With these assembly conditions, we find for MB′–DNA and NB–DNA film densities of 1.3–3.2 and 0.8–1.6 pmol/cm2, respectively.
The percent of signal remaining after incorporation of a CA mismatch was then compared as a function of film density in both low salt and spermidine buffer (Figure 5). When the percent signal remaining is plotted against the film density, a roughly linear correlation is observed. The mismatch signal attenuation improves with increasing density. This trend for MB′–DNA is present in both low salt buffer and spermidine buffer.
Figure 5.
Dependence of mismatch signal attenuation on film density for both MB′–DNA (black) and NB–DNA (gray). Percent MB′–DNA signal remaining due to the incorporation of a CA mismatch was determined in both low salt buffer (×) and spermidine buffer (squares).
The dependence of the mismatch signal attenuation on the film density suggests that there might be a contribution of the signal that originates from direct reduction at the electrode surface. DNA assembled under low density conditions (without MgCl2) has a higher surface accessibility compared to DNA assembled under high density conditions (with MgCl2), thus increasing the possibility of direct surface reduction.
Variation in Assembly Conditions: Backfilling Agent
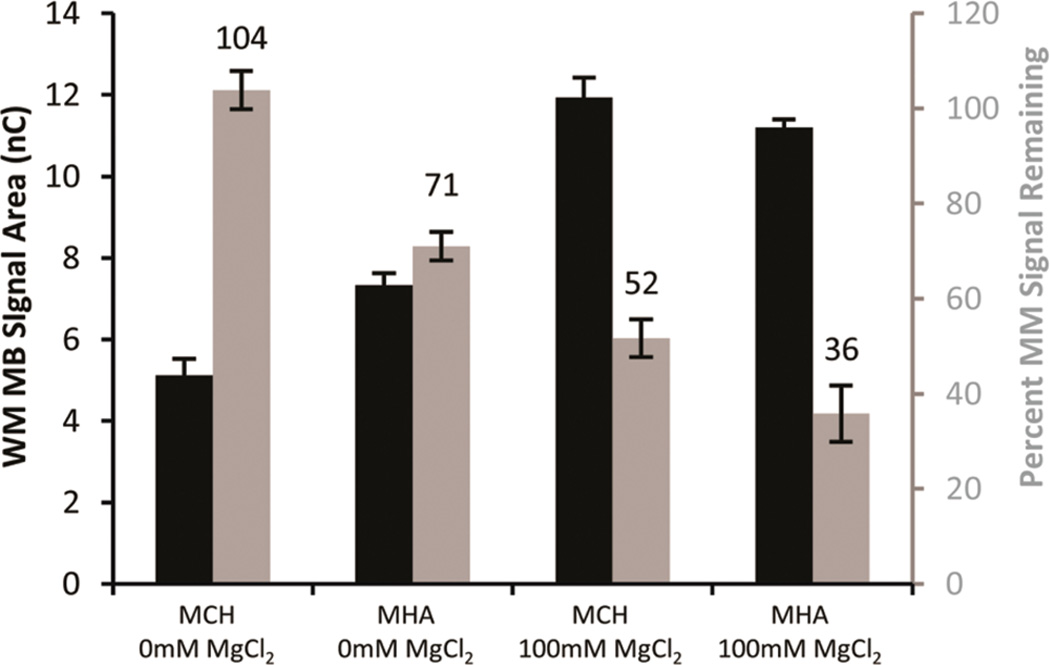
In order to gain further insight, the backfilling agent utilized was varied. Backfilling is used to decrease the background noise due to oxygen and to minimize nonspecific interactions between the DNA and the gold surface. 6-Mercaptohexanol (MCH) is the most common backfilling agent for DNA-modified electrodes. 6-Mercaptohexanoic acid (MHA) offers an alternative backfilling agent with a more negative headgroup. The degree of signal attenuation obtained, in spermidine buffer, was compared for MCH and MHA for electrodes assembled with or without 100 mM MgCl2 (Figure 6).
Figure 6.
Dependence on assembly conditions of the percent mismatch signal remaining for MB′–DNA. Well-matched signal area (black) and the percent signal remaining (gray) were from 12 electrodes averaged across three chips. The backfilling agent (1 mM MCH or MHA) and concentration of MgCl2 are indicated. Scans were acquired in spermidine buffer.
At either assembly concentration of MgCl2, switching from MCH to MHA improves the signal attenuation observed due to mismatch incorporation. When electrodes were assembled with 100 mM MgCl2 and scanned in spermidine buffer, the percent mismatch signal remaining improved from 52 ± 4% to 36 ± 6% by switching from MCH to MHA (Figure 6). Increasing the negative charge of the surface, through backfilling with MHA, increases the barrier between MB′–DNA and the electrode surface. The direct correlation between decreased surface accessibility and increased mismatch signal attenuation provides further evidence that, depending on assembly conditions, the signal observed from MB′–DNA may be generated by two different mechanisms: DNA CT and direct surface reduction.
Kinetics of MB′–DNA Reduction
The rates of electron transfer to the redox reporter for both MB′–DNA and NB–DNA were estimated under various assembly conditions, passivated with MCH, by measuring the scan rate dependence of the peak reduction potential and then applying the Laviron analysis.45 The Laviron analysis used to determine the rates of electron transfer has been routinely employed for rate determinations in these systems.12,21 Consistently, we find for DNA-mediated electrochemistry that tunneling through the linker is rate-determining and the rate is slower than with direct reduction. The same analysis was applied here to compare reduction of the reporter by DNA CT to direct surface reduction. Electrodes assembled with only single stranded (ss) MB′–DNA and NB–DNA were used to estimate the rate of electron transfer by the direct surface reduction mechanism. In this case, the complementary thiol-modified strand was omitted during assembly, ensuring that there could be no DNA-mediated contribution to the observed signal; ssDNA does not efficiently conduct charge and has a high affinity for the gold surface. For comparison, the rates of electron transfer for the DNA-mediated reduction of MB′–DNA and NB–DNA were determined under conditions where the signal attenuation due to an incorporated mismatch was maximal (double-stranded (ds) DNA assembled with 100 mM MgCl2).
It has been previously established that the rate for ssNB–DNA is 10–30-fold faster than the rate of electron transfer in dsNB–DNA, assembled with 100 mM MgCl2 and acquired in spermidine buffer.12 The rate for dsDNA is limited by tunneling through the C6-alkane linkage to the surface.12 This previous result for NB–DNA was reproduced in this study (Table 1). Furthermore, the rate of electron transfer for dsMB′–DNA, with 100 mM MgCl2, is 20-fold slower than ssMB′–DNA in both spermidine and low salt buffers (Table 1). This result confirms that dsMB′–DNA is reduced via DNA CT when assembled with 100 mM MgCl2.
Table 1.
Electron-Transfer Rate Constants of NB–DNA (kNB–DNA) and MB′–DNA (kMB′–DNA) as Single-Stranded DNA (ssDNA) and Double-Stranded DNA (dsDNA) under Different Running Conditions
| ssDNAc | dsDNAc (0 mM MgCl2) | dsDNAc (100 mM MgCl2) | ||||
|---|---|---|---|---|---|---|
| log(kMB′–DNA/s−1)a | log(kNB–DNA/s−1)a | log(kMB′–DNA/s−1)a | log(kNB–DNA/s−1)a | log(kMB′–DNA/s−1)a | log(kNB–DNA/s−1)a | |
| spermidine bufferb | 1.6 | 1.6 | 0.5 and 1.6e | 0.3 | 0.5 | 0.6 |
| low salt bufferb | 1.2 | 1.6 | −0.2 and 1.6e | N/Ad | 0.4 | N/Ad |
The electron transfer rates were determined by applying Laviron analysis to CV data acquired at scan rates ranging from 50 mV/s to 13 V/s. The uncertainties are ~10% of the log(k) values.45
Low salt buffer is 5.0 mM phosphate, 50 mM NaCl, and pH 7. Spermidine buffer is 5.0 mM phosphate, 50 mM NaCl, 4 mM MgCl2, 4 mM spermidine, 50 µM EDTA, 10% glycerol, and pH 7.
Assembled overnight with 25 µM of ssDNA (probe strand lacking thiol complement) or dsDNA, with or without 100 mM MgCl2.
The rate for double-stranded NB–DNA could not be determined in low salt buffer as the signals were too small.
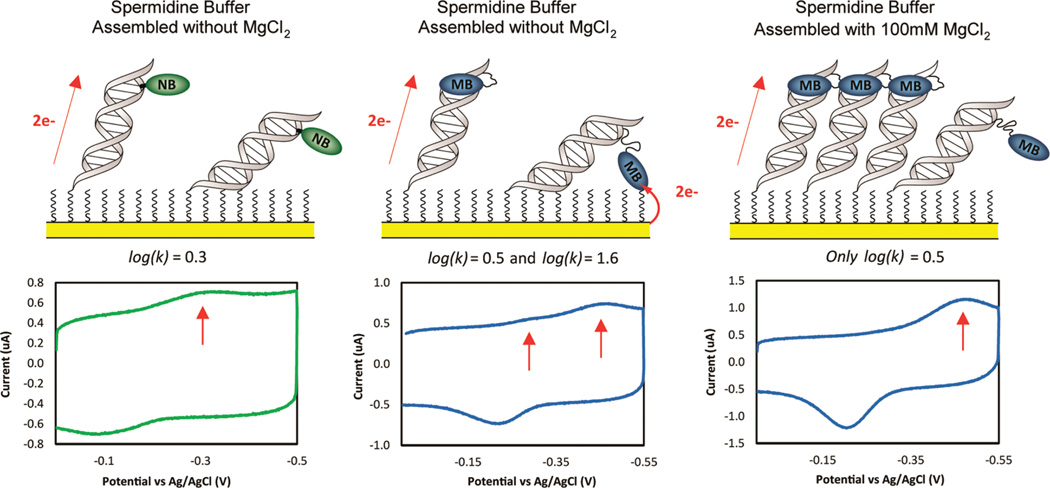
Two distinct reductive peaks were observed for MB′–DNA when assembled without MgCl2 (Figure 7).
The rates of electron transfer were then examined under conditions where DNA CT is not the sole mechanism for MB′–DNA reduction, evidenced by reduced signal attenuation from a CA mismatch (dsDNA without MgCl2). At fast scan rates (5 V/s) for dsMB′–DNA, two reductive peaks are resolved with a 15-fold rate differential in both spermidine and low salt buffer (Figure 7 and Table 1). These data indicate that rates of electron transfer for these two modes of reduction correspond with direct surface reduction and DNA-mediated reduction of MB′–DNA. Well-matched and mismatched dsMB′–DNA were compared at 5 V/s, and signal attenuation due to mismatch incorporation was observed only in the peak with a slower rate of electron transfer, consistent with it being a DNA-mediated process (data not shown). Alternatively, for dsNB–DNA only a single peak is observed in spermidine buffer with a rate that is 20-fold slower than ssNB–DNA. This observation suggests that while dsNB–DNA is only capable of being reduced via DNA CT, dsMB′–DNA may be reduced by either a DNA-mediated pathway or direct surface reduction. The ability of MB′–DNA to be reduced directly at the surface is likely due to the flexibility of the linkage through which MB′ is covalently tethered (Figure 7).
Figure 7.
Top: schematic illustration of reporter reduction mechanisms. Bottom: CV at 5 V/s (bottom) for NB–DNA (left) and MB′–DNA (center) assembled without MgCl2 and MB′–DNA assembled with 100 mM MgCl2 (right). Electrodes were backfilled with mercaptohexanol and scanned in spermidine buffer. The red arrows indicate the peaks quantified in order to determine the rate of the various processes for reporter reduction.
DISCUSSION
In this work, MB′ is covalently tethered to DNA with a flexible C12 alkyl linkage to a modified uracil. MB′–DNA produces a reversible redox couple via DNA CT. Incorporation of a single CA mismatch within a 17-mer can attenuate the mismatch signal to 7 ± 2% of the well-matched signal. The degree to which the mismatch signal is attenuated was found to be highly dependent on assembly conditions. Under conditions where DNA CT is the sole available mechanism for MB′ reduction, a 10-fold improvement in the mismatch sensitivity is obtained, when compared to previously utilized nonamplified DNA-mediated platforms.12,19 MB′–DNA was found also to be capable of being reduced directly by the surface of the electrode. The extent to which this mechanism contributes to the observed signal was found to be directly influenced by assembly conditions.
Therefore, MB′–DNA can both report on DNA CT through the intercalation of MB′ into the base stack and report on the surface accessibility of the reporter through direct surface reduction of MB′. We can discern which mechanism of MB′ reduction is dominating under any given assembly condition by examining the signal sensitivity to π-stack perturbations or by examining the rate of electron transfer.
The fact that assembly conditions can alter which mechanism is dominant for MB′ reduction has relevance in the further development of DNA-based biosensors. Sensors using direct surface reduction typically function through the formation of duplex DNA to attenuate the observed signal by increasing the separation between MB′ and the electrode.22–32 Our work shows that upon duplex formation a new pathway for dsMB′–DNA reduction has been introduced, DNA CT, which can contribute to the residual observed signal.
Recent work performed to optimize these direct surface reduction-based platforms has demonstrated that, upon duplex formation, no signal suppression is observed at slow scan rates (100 mV/s), and the duplex signal yielded a rate of electron transfer slower than that of the nonduplex DNA structure.23 In order to optimize these devices, the scan rates were increased to minimize the contributions from the dsMB′–DNA.23 The trends observed in this work are consistent with the work presented here and demonstrate that the DNA-mediated reduction of duplex MB′–DNA can significantly contribute to the observed signal.
It is clear that both direct reduction and DNA-mediated reduction need to be considered as important mechanisms in DNA-modified electrodes and that MB′–DNA is a viable reporter for both systems. Furthering our understanding of the underlying mechanism for the reduction of dsMB′–DNA is essential for exploiting both DNA CT and direct surface reduction based biosensors to their full extent. Only when both mechanisms are considered can the highest overall sensitivity be achieved.
ACKNOWLEDGMENTS
This research was supported by the National Institute of Health (GM61077). The authors thank N. Muren for discussions and her contributions in fabricating the multiplexed chips. This work was completed in part in the Caltech Micro Nano Fabrication Laboratory.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Murphy CJ, Arkin MR, Jenkins Y, Ghatlia ND, Bossmann SH, Turro NJ, Barton JK. Long Range Photoinduced Electron Transfer through a DNA Helix. Science. 1993;262:1025–1029. doi: 10.1126/science.7802858. [DOI] [PubMed] [Google Scholar]
- 2.Holmlin ER, Dandliker PJ, Barton JK. Charge Transfer Through the DNA Base Stack. Angew. Chem., Int. Ed. 1997;36:2714–2730. [Google Scholar]
- 3.Genereux JC, Boal AK, Barton JK. DNA-Mediated Charge Transport in Redox Sensing and Signaling. J. Am. Chem. Soc. 2010;132:891–905. doi: 10.1021/ja907669c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuster GB, editor. Topics in Current Chemistry. Vol. 237. Berlin: Springer-Verlag; 2004. Long Range Charge Transfer in DNA II; p. 103. [Google Scholar]
- 5.Wagenknecht HA, editor. Charge Transfer in DNA: From Mechanism to Application. Weiheim: Wiley-VCH Verlag GmbH & Co KGaA; 2005. [Google Scholar]
- 6.Genereux JG, Barton JK. Mechanisms for DNA Charge Transport. Chem. Rev. 2010;110:1642–1662. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall DB, Holmlin RE, Barton JK. Oxidative DNA Damage through Long Range Electron Transfer. Nature. 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 8.Kelley SO, Barton JK. Electron Transfer Between Bases in Double Helical DNA. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 9.Delaney S, Barton JK. Long-Range DNA Charge Transport. J. Org. Chem. 2003;68:6475–6483. doi: 10.1021/jo030095y. [DOI] [PubMed] [Google Scholar]
- 10.Kelley SO, Holmlin RE, Stemp EDA, Barton JK. Photoinduced Electron Transfer in Ethidium-Modified DNA Duplexes: Dependence on Distance and Base Stacking. J. Am. Chem. Soc. 1997;119:9861–9870. [Google Scholar]
- 11.Gorodetsky A, Buzzeo MC, Barton JK. DNA-mediated Electrochemistry. Bioconjugate Chem. 2008;19:2285–2296. doi: 10.1021/bc8003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slinker J, Muren N, Renfrew S, Barton JK. DNA charge transport over 34 nm. Nat. Chem. 2011;3:228–233. doi: 10.1038/nchem.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelley SO, Jackson NM, Hill MG, Barton JK. Long Range Electron Transfer Through DNA Films. Angew. Chem., Int. Ed. 1999;38:941. doi: 10.1002/(SICI)1521-3773(19990401)38:7<941::AID-ANIE941>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 14.Boon EM, Salas JE, Barton JK. An Electrical Probe of Protein-DNA Interactions on DNA-Modified Surfaces. Nat. Biotechnol. 2002;20:282–286. doi: 10.1038/nbt0302-282. [DOI] [PubMed] [Google Scholar]
- 15.Kelley SO, Boon EM, Barton JK, Jackson NM, Hill MG. Single-Base Mismatch Detection Based on Charge Transduction Through DNA. Nucleic Acids Res. 1999;27:4830–4837. doi: 10.1093/nar/27.24.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boon EM, Ceres DM, Drummond TG, Hill MG, Barton JK. Mutation Detection by Electrocatalysis at DNA-Modified Electrodes. Nat. Biotechnol. 2000;18:1096–1100. doi: 10.1038/80301. [DOI] [PubMed] [Google Scholar]
- 17.Gorodetsky A, Green O, Yavin E, Barton JK. Coupling into the Base Pair Stack is Necessary for DNA-mediated Electrochemistry. Bioconjugate Chem. 2007;18:1434–1441. doi: 10.1021/bc0700483. [DOI] [PubMed] [Google Scholar]
- 18.Kelley SO, Barton JK, Jackson N, Hill MG. Electrochemistry of Methylene Blue bound to a DNA-Modified Electrode. Bioconjugate Chem. 1997;8:31–37. doi: 10.1021/bc960070o. [DOI] [PubMed] [Google Scholar]
- 19.Slinker J, Muren N, Gorodetsky A, Barton JK. Multiplexed DNA-Modified Electrodes. J. Am. Chem. Soc. 2010;132:2769–2774. doi: 10.1021/ja909915m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boon E, Jackson N, Wightman M, Kelley S, Hill M, Barton JK. Intercalative Stacking: A Critical Feature of DNA Charge-Transport Electrochemistry. J. Phys. Chem. B. 2003;107:11805–11812. [Google Scholar]
- 21.Drummond TG, Hill MG, Barton JK. Electron Transfer Rates in DNA Films as a Function of Tether Length. J. Am. Chem. Soc. 2004;126:15010–15011. doi: 10.1021/ja044910i. [DOI] [PubMed] [Google Scholar]
- 22.Kang D, Zuo X, Yang R, Xia F, Plaxco K, White R. Comparing the Properties of Electrochemical-Based DNA Sensors Employing Different Redox Tags. Anal. Chem. 2009;81:9109–9113. doi: 10.1021/ac901811n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W, Lai RY. Comparison of the Stem-Loop and Linear Probe-Based Electrochemical DNA Sensors by Alternating Current Voltammetry and Cyclic Voltammetry. Langmuir. 2011;27:14669–14677. doi: 10.1021/la203015v. [DOI] [PubMed] [Google Scholar]
- 24.Xiang Y, Qian X, Chen Y, Zhang Y, Chai Y, Yuan R. A reagentless and disposable electronic genosensor: from multiplexed analysis to molecular logic gates. Chem. Commun. 2011;47:2080–2082. doi: 10.1039/c0cc04350h. [DOI] [PubMed] [Google Scholar]
- 25.Yang W, Lai RY. Effect of diluent chain length on the performance of the electrochemical DNA sensor at elevated temperatures. Analyst. 2011;136:134–139. doi: 10.1039/c0an00644k. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, Yang W, Lai RY. A folding-based electrochemical aptasensor for detection of vascular endothelial growth factor in human whole blood. Biosens. Bioelectron. 2011;26:2442–2447. doi: 10.1016/j.bios.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 27.Yang W, Lai RY. Integration of two different sensing modes in an electrochemical DNA sensor for approximation of target mismatch location. Electrochem. Commun. 2011;13:989–992. [Google Scholar]
- 28.Farjami E, Clima L, Gothelf K, Ferapontova EE. “Off-On” Electrochemical Hairpin-DNA-Based Genosensor for Cancer Diagnostics. Anal. Chem. 2011;83:1594–1602. doi: 10.1021/ac1032929. [DOI] [PubMed] [Google Scholar]
- 29.Xiao Y, Lai R, Plaxco K. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat. Protocols. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]
- 30.Xiao Y, Lou X, Uzawa T, Plakos K, Plaxco K, Soh HT. An Electrochemical Sensor for Single Nucleotide Polymorphism Detection in Serum Based on a Triple-Stem DNA Probe. J. Am. Chem. Soc. 2009;131:15311–15316. doi: 10.1021/ja905068s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lubin A, Vander Stoep Hunt B, White R, Plaxco K. Effects of Probe Length, Probe Geometry, and Redox-Tag Placement on the Performance of the Electrochemical E-DNA Sensor. Anal. Chem. 2009;81:2150–2158. doi: 10.1021/ac802317k. [DOI] [PubMed] [Google Scholar]
- 32.Cash K, Heeger A, Plaxco K, Xiao Y. Optimization of a Reusable, DNA Pseudoknot-Based Electrochemical Sensor for Sequence-Specific DNA Detection in Blood Serum. Anal. Chem. 2009;81:656–661. doi: 10.1021/ac802011d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ricci F, Bonham A, Mason A, Reich N, Plaxco K. Reagentless, Electrochemical Approach for the Specific Detection of Double-and Single-Stranded DNA Binding Proteins. Anal. Chem. 2009;81:1608–1614. doi: 10.1021/ac802365x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuo W, Song S, Zhang J, Pan D, Wang L, Fan C. A Target-Responsive Electrochemical Aptamer Switch (TREAS) for Reagentless Detection of Nanomolar ATP. J. Am. Chem. Soc. 2007;129:1042–1043. doi: 10.1021/ja067024b. [DOI] [PubMed] [Google Scholar]
- 35.Di Giusto DA, Wlassoff WA, Giesebrecht S, Gooding JJ, King GC. Enzymatic Synthesis of Redox-Labeled RNA and Dual-Potential Detection at DNA-modified Electrodes. Angew. Chem., Int. Ed. 2004;43:2809–2812. doi: 10.1002/anie.200352977. [DOI] [PubMed] [Google Scholar]
- 36.Di Giusto DA, Wlassoff WA, Giesebrecht S, Gooding JJ, King GC. Multipotential Electrochemical Detection of Primer Extension Reactions on DNA Self-Assembled Monolayers. J. Am. Chem. Soc. 2004;126:4120–4121. doi: 10.1021/ja0319036. [DOI] [PubMed] [Google Scholar]
- 37.Anne A, Bouchardon A, Moiroux J. 3′-Ferrocene-Labeled Oligonucleotide Chains End-Tethered to Gold Electrode Surfaces: Novel Model Systems for Exploring Flexibility of Short DNA Using Cyclic Voltammetry. J. Am. Chem. Soc. 2003;125:1112–1113. doi: 10.1021/ja028640k. [DOI] [PubMed] [Google Scholar]
- 38.Boon EM, Barton JK, Bhaghat V, Nerissian M, Wang W, Hill MG. Reduction of Ferricyanide by Methylene Blue at a DNA-Modified Rotating-Disk Electrode. Langmuir. 2003;19:9255–9259. [Google Scholar]
- 39.Leng F, Savkur R, Fokt I, Przewloka T, RPiebe W, Chaires JB. Base Specific and Regioselective Chemical Cross-Linking of Daunorubicin to DNA. J. Am. Chem. Soc. 1996;118:4732. [Google Scholar]
- 40.Wang AHJ, Gao YG, Liaw YC, Li YK. Formaldehyde crosslinks daunorubicin and DNA efficiently: HPLC and x-ray diffraction studies. Biochemistry. 1991;30:3812. doi: 10.1021/bi00230a002. [DOI] [PubMed] [Google Scholar]
- 41.Wagner SJ, Skripchenko A, Robinette D, Foley JW, Cincotta L. Factors Affecting Virus Photoinactivation by a Series of Phenothiazine Dyes. Photochem. Photobiol. 1998;67:343–349. [PubMed] [Google Scholar]
- 42.Chen H, Herkstroeter WG, Perlstein J, Law KY, Whitten DG. Aggregation of a Surfactant Squaraine in Langmuir-Blodgett Films, Solids, and Solution. J. Phys. Chem. 1994;98:5138–5146. [Google Scholar]
- 43.Pedersen DS, Rosenbohm C. Dry Column Vacuum Chromatography. Synthesis. 2001;16:2431–2434. [Google Scholar]
- 44.The reduction potential of NB is shifted positive by 200mV due to the direct conjugation of NB to the nucleobase [Google Scholar]
- 45.Laviron EJ. General expression of the linear potential sweep voltammagram in the case of diffusionless electrochemical systems. J. Electroanal. Chem. 1979;101:19–28. [Google Scholar]