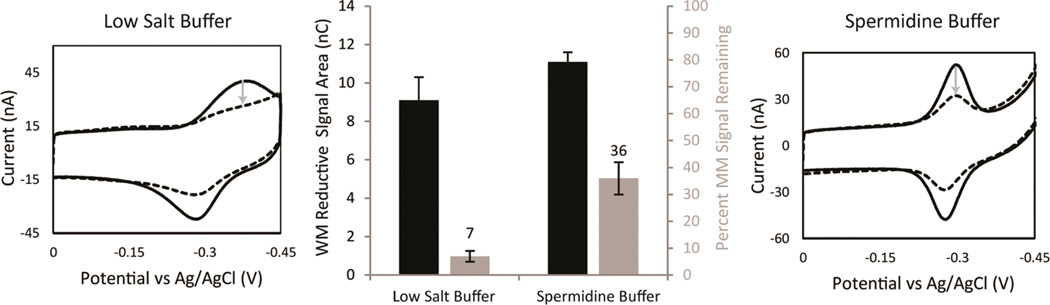
Figure 4.
Optimization of mismatch discrimination depending on the running buffer. Both WM MB′–DNA (solid) and MM10 MB′–DNA (dotted) modified electrodes were assembled with 100 mM MgCl2, passivated with MHA, and scanned in both a low salt buffer (5.0 mM phosphate, 50 mM NaCl, and pH 7) (right) and a spermidine buffer (5.0 mM phosphate, 50 mM NaCl, 4 mM MgCl2, 4 mM spermidine, 50 µM EDTA, 10% glycerol, and pH 7) (left). The areas of the reductive peaks were used to quantify the reductive signal size (black) and determine the percent signal remaining ([MM]/[WM] × 100) (gray) from the incorporation of a single mismatch. The gray arrows denote the decrease in signal area with the optimal percent signal attenuation being in low salt buffer. The errors denoted were determined by the standard deviation from four electrodes averages, for each sequence of DNA, across three chips.