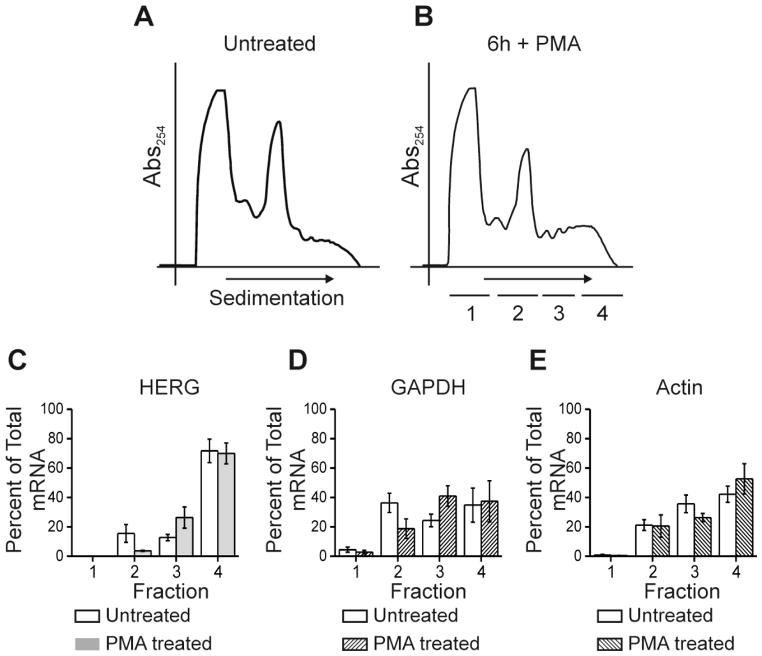
Figure 5. PMA treatment does not alter HERG mRNA association with polysomes.
Parts A and B show polyribosomal profiles for untreated and 6-hour PMA treated conditions. The y-axis shows absorbance at 254 nm plotted against increasing gradient density where heaviest components will sediment to the right. The underlined segments 1–4 show where fractions were collected for subsequent qRT-PCR. Parts C-E show summary data for results of qRT-PCR. The y-axis values represent percent of total RNA that is comprised by (C) HERG, (D) GAPDH, and (E) Actin, respectively per fraction. Open bars show the untreated condition while shaded bars show the cAMP treated condition. Differences between treated and untreated conditions were not significant, n=4.