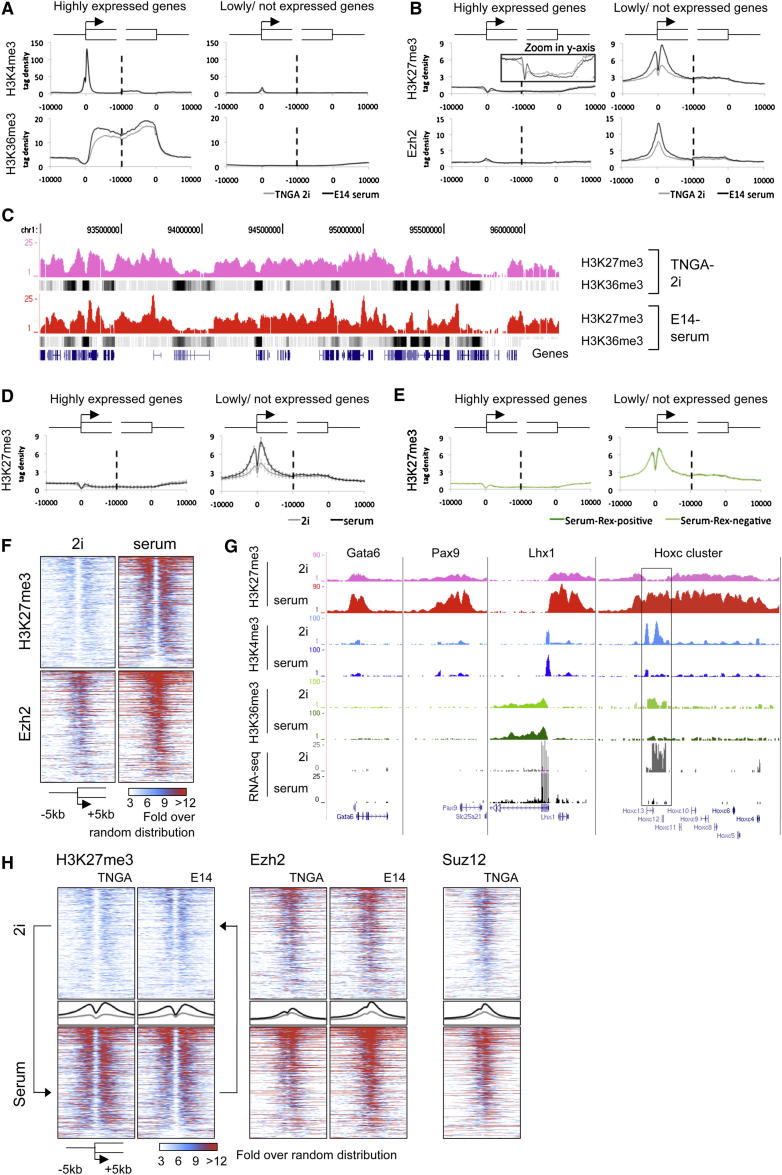
Figure 3.
H3K27me3 Is Greatly Diminished at Promoters of Silent Genes and at Hox Clusters in 2i
(A) Average H3K4me3 and H3K36me3 profiles of the 2,000 most active (left plots) and 2,000 silent genes (right plots) from −10kb to +10kb at the transcription start site and the transcription stop. The negative control (genomic DNA) is shown in Figure S3B.
(B) Average profiles of H3K27me3 and Ezh2 associated with silencing as in (A). Figure S3G shows a biological replicate analysis for the H3K27me3 profiles of TNGA 2i ES cells.
(C) Typical example of H3K27me3 and H3K36me3 (dense setting) profiles.
(D) H3K27me3 profiling for three different cell lines maintained and derived in either 2i or serum, as in (B). H3K27me3 profiles were generated for TNGA, NOD_male, and NOD_female ES cells in 2i and E14, HM1, and RGD2 ES cells in serum.
(E) As in (B); H3K27me3 profiling in Rex1-positive and Rex1-negative serum ES cells.
(F) H3K27me3 and Ezh2 intensity plots of all promoters containing H3K27me3 at >3-fold over random distribution in at least one of the conditions, TNGA-2i or E14-serum; 3,870 promoters are depicted in rows on the y axis.
(G) Typical examples showing H3K27me3 reduction in 2i as compared to serum.
(H) As in (F); promoter profiles for 2i ES cells adapted to serum and vice versa. Average profiles are plotted in the middle (gray: 2i; black: serum).