Figure S3.
H3K27me3 Is Greatly Diminished at Promoters of Silent Genes and at Hox Clusters in ES Cells in 2i, Related to Figure 3
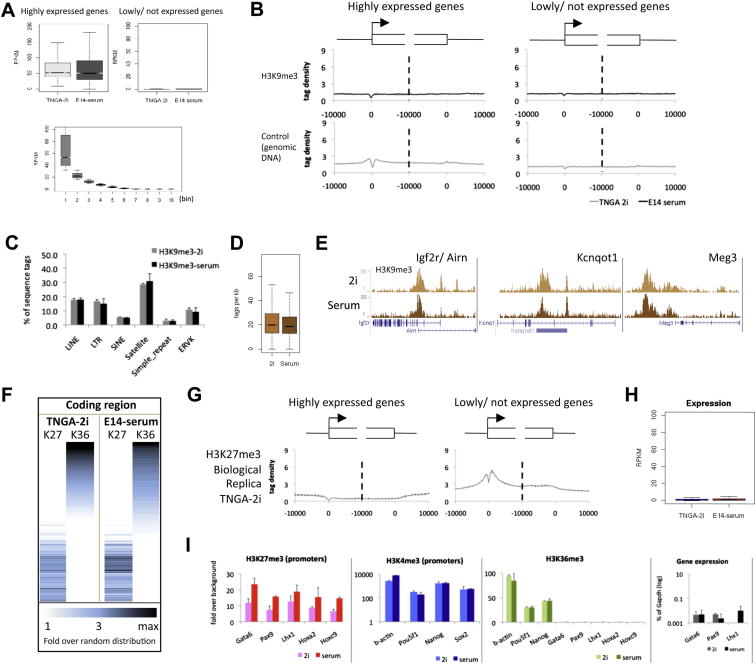
(A) Expression levels of the two groups of genes shown in Figure 3A. For comparative purposes, the expression level of all genes equally divided into ten bins according to expression level is shown at the bottom.
(B) Similar to Figures 3A and 3B: Average H3K9me3 profiles over active and inactive genes, as well as the negative control (genomic DNA derived from the chromatin input).
(C) Percentage of H3K9me3 tags present in the major repeat categories.
(D) Quantification of the H3K9me3 enriched genomic loci, as determined by Yuan et al. (2009), in 2i and serum.
(E) Typical examples of H3K9me3, enrichment over three imprinted genes.
(F) H3K27me3 (“K27”) and H3K36me3 (“K36”) intensity plots for the genomic coding regions of all genes (ranked by TNGA-2i H3K36me3 values), showing that H3K27me3 and H3K36me3 are mutual exclusive.
(G) Similar to Figure 3B: H3K27me3 profiling of biological replica of TNGA in 2i.
(H) Expression of the genes shown in Figure 3F. See Figure S3A (bottom) for a binned overview for the expression level of all genes.
(I) Validation of the ChIP-seq and RNA-seq profiles by ChIP-qPCR or RT-qPCR, respectively.