Abstract
HOXA9 plays a critical role in both normal hematopoiesis and leukemogenesis, particularly in the development and maintenance of mixed lineage leukemia (MLL)-rearranged leukemia. Through reverse transcription polymerase chain reaction (RT-PCR) analysis of HOXA9 transcripts in human leukemia and normal bone marrow samples, we identified a truncated isoform of HOXA9, namely HOXA9T, and found that both HOXA9T and canonical HOXA9 were highly expressed in leukemia cell lines bearing MLL rearrangements, relative to human normal bone marrow cells or other subtypes of leukemia cells. A frameshift in HOXA9T in exon I causes a premature stop codon upstream of the PBX binding domain and the homeodomain, which leads to the generation of a non-homeodomain-containing protein. Unlike the canonical HOXA9, HOXA9T alone cannot transform normal bone marrow progenitor cells. Moreover, HOXA9T cannot cooperate with MEIS1 to transform cells, despite the presence of a MEIS1-binding domain. Remarkably, although the truncated isoforms of many proteins function as dominant-negative competitors or inhibitors of their full-length counterparts, this is not the case for HOXA9T; instead, HOXA9T synergized with HOXA9 in transforming mouse normal bone marrow progenitor cells through promoting self-renewal and proliferation of the cells. Collectively, our data indicate that both truncated and full-length forms of HOXA9 are highly expressed in human MLL-rearranged leukemia, and the truncated isoform of HOXA9 might also play an oncogenic role by cooperating with canonical HOXA9 in cell transformation and leukemogenesis.
Keywords: HOXA9, HOXA9T, isoforms, leukemia
INTRODUCTION
HOX genes are a highly conserved subgroup of the homeobox superfamily which plays an essential role during embryogenesis and oncogenesis [1; 2]. In mammals, 39 HOX genes are clustered in four different chromosomal loci, designated as clusters A, B, C and D [3]. HOX genes encode transcription factors and bind to DNA through a highly conserved 183 nucleotide sequence called the homeodomain. HOX proteins can function as monomers or homodimers to drive the transcription of downstream targets directly, and as heterodimers or heterotrimers with members of the three amino acid loop extension (TALE) family of cofactors, such as MEIS or PBX [4; 5; 6; 7].
HOXA9 is a well-studied member of the HOXA cluster, and it plays a significant role in both normal hematopoiesis and leukemogenesis. Hoxa9 deficient mice showed impaired hematopoietic development [8]. In humans, HOXA9 was found to be highly expressed and required in some subtypes of leukemia, particularly mixed lineage leukemia (MLL)-rearranged leukemia [9; 10; 11; 12]. The aberrant overexpression of HOXA9 is thought to be associated with poor survival of acute myeloid leukemia (AML) patients [11; 13; 14; 15]. Moreover, enforced coexpression of Hoxa9 and its co-factor Meis1 in mouse bone marrow cells leads to rapid AML development [7; 16], and they are able to replace the leukemogenic activity of MLL-ENL [17].
The canonical HOXA9 (hereafter called HOXA9 for simplicity) transcript contains two exons: exon I and the homeodomain-containing exon II [18; 19]. In mice, an alternatively spliced transcript (namely Hoxa9T) which resulted in a frameshift in exon I leading to production of a truncated protein lacking the homeobox domain was identified a decade ago [20; 21]. Hoxa9T is significantly expressed in different mouse embryonic tissue. When ectopically expressed in Hela cells, Hoxa9 was strictly targeted to the nucleus, while the Hoxa9T was found both in the nucleus and the cytoplasm. It was also found that both isoforms were able to interact with coactivator CREB-binding protein (CBP), but only Hoxa9 was able to interact with Meis1, Meis2 and Pbx1b [20]. However, the presence of the HOXA9T in humans has only been reported in an endometrial adenocarcinoma cell line [20]; HOXA9T has not been reported in normal human cells and its function remains elusive.
Due to the aberrant overexpression and the significant oncogenic role of HOXA9 in leukemia, we determined the presence of HOXA9 isoforms in human leukemia cell lines and normal bone marrow mononuclear cells by use of the RT-PCR assay, followed by sequencing. We identified two alternatively spliced transcripts of human HOXA9 genes designated HOXA9 and HOXA9T that encode for a homeodomain- and a non-homeodomain-containing protein, respectively. Both were highly expressed in human MLL-rearranged leukemia cell lines compare to normal bone marrow mononuclear cells or other subtypes of leukemia cells. In addition, we employed mouse bone marrow progenitor cell colony-forming and replating assays to study the function of the two isoforms. We found that HOXA9T alone or together with MEIS1 could not transform normal bone marrow progenitor cells. Nonetheless, HOXA9T is not a dominant negative competitor of HOXA9; instead, forced expression of HOXA9T significantly enhanced the ability of HOXA9 to transform mouse bone marrow progenitor cells.
MATERIALS AND METHODS
Cell culture and normal control samples
MONOMAC6 (containing an MLL-AF9 fusion gene) cells were growing in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) (Invitrogen), 2 mM L-glutamine, nonessential amino acid, 1mM sodium pyruvate and 9 μg/ml human insulin. THP-1 (also containing an MLL-AF9 fusion gene) was maintained in RPMI 1640 containing 1% penincillin-streptomycin, 0.05 mM 2-mercaptoethanol and 10% FBS. KOCL-48/t(4;11), KASUMI-1/t(8;21) and NB-4/t(15;17) were maintained in RPMI 1640 containing 1% penincillin-streptomycin, 0.05 mM 2-mercaptoethanol and 10% FBS. HEK293T and Rat1a cells were cultured in Dulbecco modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% penincillin-streptomycin (Invitrogen). The mononuclear cell (MNC) samples were isolated from normal BM cells purchased from AllCells, LLC. MNC cells were isolated using NycoPrep 1.077A (Axis-Shield, Oslo, Norway) according to the manufacturer's manual.
RNA preparation, PCR and DNA sequencing
The total RNA of mononuclear cells and cell lines was isolated by use of miRNeasy Mini Kit (Qiagen). One step reverse transcription-polymerase chain reaction (RT-PCR) was performed with the primers: HOXA9-forward 5'-atagaattcatggccaccactggggc-3', HOXA9-reverse 5'-accctcgagtcactcgtcttttgctc-3' using one-step RT-PCR kit (Qiagen). The PCR condition is reverse transcription at 50°C for 30min, initial PCR activation step at 95°C for 15min, 30 cycles of 3-step cycling with 94°C for 30 seconds, 60°C for 30 seconds and 72°C for 1min, and final extension at 72°C for 10min. RT-PCR product was run in an agarose gel. DNA was recovered from the gel with QIAquick Gel Extraction Kit (Qiagen) and was sequenced with primer 5'-atagaattcatggccaccactggggc-3'.
Retroviral constructs cloning
HOXA9 and HOXA9T were PCR-amplified from human normal bone marrow mononuclear cells, and were then cloned into MSCV-PIG vector that contains a PGK-puromycin-IRES-GFP (PIG)[22]. HOXA9 and MEIS1 were PCR amplified from human normal bone marrow mononuclear cells, and were cloned into MSCVneo (Clontech, Mountain View, CA). The primers are shown in Table 1.
Table 1.
Primer sequence for retroviral construct cloning.
| Name | Primer sequence | Restriction enzyme |
|---|---|---|
| HOXA9-F-MSCV-PIG | atactcgagatggccaccactggggc | XhoI |
| HOXA9-R-MSCV-PIG | accgaattctcactcgtcttttgctc | EcoRI |
| HOXA9-F-MSCVneo | atagaattcatggccaccactggggc | EcoRI |
| HOXA9-R-MSCVneo | accctcgagtcactcgtcttttgctc | XhoI |
| MEIS1-F-MSCVneo | atagaattcatggcgcaaaggtac | EcoRI |
| MEIS1-R-MSCVneo | ggcctcgagtagatgaaggttaca | XhoI |
Retrovirus preparation
As described previously [23; 24], retrovirus for each construct was produced in 293T cells by co-transfecting the retroviral constructs and pCL-Eco packaging vector (IMGENEX, San Diego, CA) using Effectene transfection reagent(Qiagen). Rat1a cells were used to determine the viral titer.
In vitro colony-forming and replating assays
Hematopoietic progenitor (i.e., lineage negative, Lin-) cells were obtained from a cohort of 4- to 6-week-old B6.SJL (CD45.1) mice five days after 5-FU treatment (150 mg/kg) using the Mouse Lineage Cell Depletion Kit (Miltenyi Biotec Inc., Auburn, CA), and were then co-transduced with retrovirus collected from 293T cells transfected with retroviral constructs through “spinoculation”[23; 24]. Then, four aliquots of 1× 104 of the transfected cells were plated into four 35 mm Nunc Petri dishes in 1.1 ml of Methocult M3230 methylcellulose medium (Stem Cell Technologies Inc., Vancouver, BC, Canada) containing 10 ng/ml each of murine recombinant IL-3, IL-6, and GM-CSF and 30 ng/ml of murine recombinant stem cell factor (R&D Systems, Minneapolis, MN), along with 1.0 mg/ml of G418 (Gibco BRL, Gaithersburg, MD) and 2.5 μg/ml of puromycin (Sigma, St. Louis, MO). Cultures were incubated at 37°C in a humidified atmosphere of 5% CO2 in air for 7 days. Then, cells from colonies in each dish were washed and collected, and replated in methylcellulose dishes every 7 days with 1× 104 cells for up to three passages.
Histopathology
Cells were collected from colony-forming and replating assays. 50,000 cells were washed twice with cold MACS Buffer and were diluted in 200 μl of the same buffer. Each sample was loaded into the appropriate wells of the cytospin, and then spun at 2000 rpm for 2 min. The cytospin slides were stained with Wright-Giemsa and were analyzed by us and by members of our hematopathology faculty.
RESULTS
Two isoforms of HOXA9 are highly expressed in MLL-rearranged leukemia cells
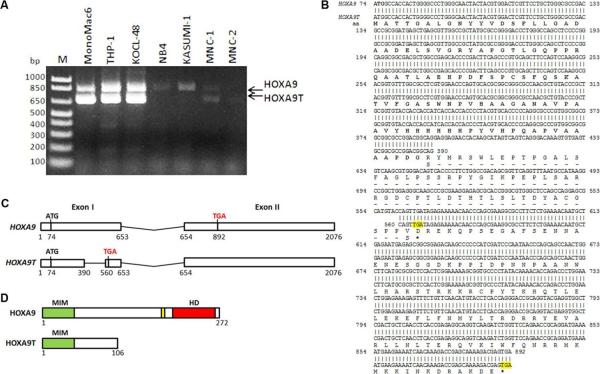
To investigate the presence and abundance of HOXA9 transcript isoforms in leukemia cell lines, we performed RT-PCR in five leukemia cell lines including MONOMAC6/t(9;11), THP-1/t(9;11), KOCL-48/t(4;11), NB-4/t(15;17) and KASUMI-1/t(8;21), along with two normal human bone marrow mononuclear cell samples. We observed that two transcripts (around 830bp and 660bp, respectively) were present in the cells and the expression level of both transcripts was much higher in MLL-rearranged leukemia cell lines, such as MONOMAC6, THP-1 and KOCL-48, compared to normal control cells or two other leukemia cell lines, NB-4 and KASUMI-1 (Fig. 1A). In order to determine the sequence of the two transcripts, we recovered the DNA fragments from the gel and sequenced them. After aligning with the NCBI nucleotide database, we found that the longer transcript was the full length HOXA9 coding region sequence (CDS), while the shorter one was an alternative splicing isoform of HOXA9 (i.e. HOXA9T) which was missing 174bp in the middle of the first exon (Figs. 1B and C). Interestingly, this isoform causes a frame shift and introduces a stop codon four base pairs after the deletion. As a result, there is no homeodomain and PBX-binding domain in the truncated HOXA9, but the MEIS-binding domain is retained (see Figs. 1B and D).
Figure 1. HOXA9 expression in leukemia cell lines, sequence and the schematic structures.
(A) Two transcripts of HOXA9 were detected in normal human bone marrow mononuclear cells (MNC) and leukemia cell lines including MONOMAC6 /t(9;11), KOCL-48 /t(4;11), THP-1 /t(9;11), NB-4/t(15;17) and KASUMI-1/t(8;21) by RT-PCR. (B) Alignment of the sequence of two isoforms of HOXA9. RT-PCR product of the two transcripts from MONOMAC6, KOCL-48, and THP-1 was sequenced after gel recovery. The sequences of HOXA9 (upper row) and HOXA9T (lower row) were aligned. The stop codes were highlighted. Amino acid (aa) sequence was also showed below the nucleotide sequence. The common amino acid sequence between HOXA9 and HOXA9T were shown in boldface. Asterisks indicate stop codons. (C) Schematic of gene structure of human HOXA9 and HOXA9T. The exons (exon I and exon II) are represented with white boxes. The nucleotide locations are indicated below the exons. Initiation (ATG) and stop codons (TGA) are also shown. (D) Schematic of protein structure of human HOXA9 and HOXA9T. MIM, MEIS1 interaction motif; HD, homeobox domain; yellow box, tryptophan consensus motif for PBX binding.
HOXA9T alone or together with MEIS1 cannot induce cell transformation
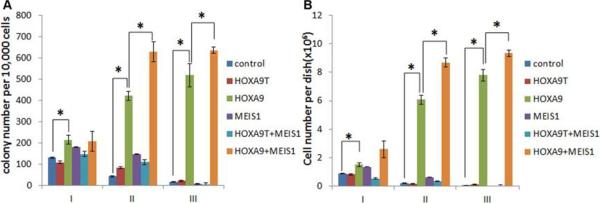
Previous studies showed that forced expression of HOXA9 alone can induce cell immortalization in vitro [25; 26]. To compare the roles of HOXA9 and HOXA9T in cell transformation, we performed in vitro colony-forming and replating assays. As expected, forced expression of HOXA9 alone in mouse normal bone marrow progenitor cells resulted in hundreds of colonies after series of replating; however, forced expression of HOXA9T alone caused only a few colonies, similar to transduction of empty vector controls (see Fig. 2A). These data suggest that unlike HOXA9, HOXA9T alone cannot transform normal bone marrow progenitor cells.
Figure 2. In vitro mouse bone marrow progenitor cell colony-forming and replating assays with HOXA9, HOXA9T and/or MEIS1.
Colony-forming and replating assays of mouse normal bone marrow progenitor cells transduced with MSCV-PIG+MSCVneo (i.e. control), HOXA9T_MSCV-PIG+MSCVneo (i.e. HOXA9T), HOXA9_MSCV-PIG+MSCVneo (i.e. HOXA9), MSCV-PIG+MEIS1_MSCVneo (i.e. MEIS1), HOXA9T_MSCV-PIG+ MEIS1_MSCVneo (i.e. HOXA9T+MEIS1) or HOXA9_MSCV-PIG+ MEIS1_MSCVneo (i.e. HOXA9+MEIS1). Duplicates were plated for each combination with 1×104 cells per dish, and every 7 days the cells were replated for up to three passages. Two independent experiments were conducted. Mean±SD values are shown. *, P<0.05, two-tailed t-test. (A) Average numbers of colonies per dish (≥ 50 cells/colony; 1×104 input cells) standard deviation of different passages (I, II and III) are shown. (B) Average cell numbers per dish and standard deviation values are shown.
Although HOXA9T does not contain the homeodomain, the MEIS1-binding domain is still retained in its N terminus [27] (Fig. 1D). Thus, we sought to investigate whether HOXA9T can cooperate with MEIS1 to induce cell transformation. As shown in Figure 2A, though forced expression of MEIS1 alone could not transform mouse bone marrow progenitor cells, co-expression of MEIS1 and HOXA9 resulted in signficantly more colonies than forced expression of each alone. However, there was no synergistic effect between HOXA9T and MEIS1, and co-expression of both did not cause many colonies either (Fig. 2A). Similar patterns were observed when we compared the average number of cells per dish in the colony-forming and replating assays (Fig. 2B). These findings indicate that although HOXA9T contains a MEIS1-binding domain, HOXA9T does not cooperate with MEIS1 to transform normal cells.
HOXA9T can enhance the cell-transformation capability of HOXA9
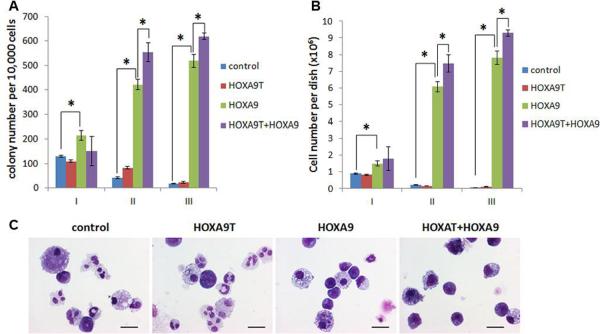
Truncated isoforms of many proteins have been shown to function as dominant-negative competitors or inhibitors of the full-length counterparts [28; 29; 30; 31; 32; 33; 34]. Thus, we sought to determine whether HOXA9T also acts as a dominant-negative antagonist of HOXA9. Surprisingly, however, in our colony-forming and replating assays, we found that co-transduction of HOXA9T and HOXA9 (i.e., HOXA9T+HOXA9) into mouse normal bone marrow progenitor cells caused significantly (P<0.05) more colonies than transduction of each alone after series of replating (see Fig. 3A). The cells number from each dish was consistant with the pattern of the colony number (Fig. 3B). To determine cell morphology, we obtained cytospin preparations and stained them with Wright-Giemsa. Cells with HOXA9T overexpression presented similar morphology as the control cells in which most of the cells differentiated into macrophages or neutrophils, while many of the cells transduced with HOXA9 displayed a very immature morphology (basophilic cytoplasm, large nucleus, evident nucleolus, lacy chromatin) (Fig. 3C). Notably, there were even a higher proportion of immature cells in the colonies containing both HOXA9T and HOXA9 compare to HOXA9 alone (Fig. 3C). Together, these results indicate that although HOXA9T alone is not a strong oncogene, it exhibits a synergistic oncogenic effect with HOXA9 to transform mouse bone marrow progenitor cells through promoting cell self-renewal and proliferation.
Figure 3. There is a synergistic effect between HOXA9T and HOXA9 in cell transformation.
Colony-forming and replating assays of mouse normal bone marrow progenitor cells transduced with MSCV-PIG+MSCVneo (i.e. control), HOXA9T_MSCV-PIG+MSCVneo (i.e. HOXA9T), HOXA9_MSCV-PIG+MSCVneo (i.e. HOXA9) or HOXA9T_MSCV-PIG+HOXA9_MSCVneo (i.e. HOXA9T+ HOXA9). Duplicates were plated for each combination with 1×104 cells per dish, and every 7 days the cells were replated for up to three passages. Two independent experiments were conducted. Mean±SD values are shown. *, P<0.05, two-tailed t-test. (A) Average numbers of colonies per dish (≥ 50 cells/colony; 1×104 input cells) standard deviation of different passages (I, II and III) are shown. (B) Average cell numbers per dish and standard deviation values are shown. (C) Morphology of colony cells of the secondary passage. Cells were stained with Wright-Giemsa. Scale bars represent 10 μm.
DISCUSSION
In the present study, we identified a truncated isoform of HOXA9, namely HOXA9T, in human hematopoietic cells and showed that both HOXA9T and HOXA9 are highly expressed in human leukemia cell lines with MLL rearrangements, such as MONOMAC6, THP-1 and KOCL-48, compare to normal bone marrow cells or the leukemia cell lines with other chromosomal translocations (Fig. 1A). Furthermore, through sequencing analysis we found that the alternative splicing isoform of HOXA9 resulted in a frameshift which lead to the generation of a truncated protein lacking the homeobox (see Fig. 1B–D).
Although the expression of this truncated isoform of Hoxa9 has been found previously in several other species [18; 19; 20; 21], little is known about the expression and function of HOXA9T in normal human cells including the hematopoietic system. The overexpression of the truncated isoform of HOXA9 in MLL-rearranged leukemia cell lines (Fig. 1A) implies that HOXA9T might also play a role to leukemogenesis in MLL-rearranged leukemia. Nonetheless, in colony-forming and replating assays, we found that unlike HOXA9, HOXA9T alone could not transform mouse bone marrow progenitor cells in vitro (see Fig. 2A). This is not surprising, because HOXA9T loses the homeobox domain which is essential for DNA binding, and thus is likely to lose the ability to drive the transcription of downstream targets directly.
It is well-known that TALE family homeobox proteins, such as MEIS1, PBX, bind with HOXA9 and increase its DNA-binding affinity and thus enhance the transcriptional activity of HOXA9 [6; 35; 36]. Even though HOXA9T lacks the homeobox and PBX-binding domains which are located in the C-terminus, MEIS1-binding domain which is in the N terminus remains [27]. However, we didn't see any synergistic or additive effects between HOXA9T and MEIS1 in cell transformation (see Fig. 2). This is consistent with previous findings that Hoxa9T does not interact significantly with Meis1, Meis2 in vitro in GST pull-down assays whereas Hoxa9 protein interacts with these partners [20].
Numerous studies showed that truncated isoforms of many proteins usually function as dominant-negative competitors or inhibitors of their full-length counterparts [28; 29; 30; 31; 32; 33; 34]. Given the fact that HOXA9T itself cannot transform cells and that both Hoxa9T and Hoxa9 are able to bind CBP with high efficiency [19; 20], one may expect that HOXA9T might function as a dominant-negative competitor or inhibitor of HOXA9 via some potential mechanism such as competing for CBP binding. However, we demonstrated that this is not the case; instead, HOXA9T exhibited a synergistic effect with HOXA9 in transforming hematopoietic progenitor cells, probably through forming heterodimers with HOXA9 or via some unknown mechanisms. Thus, our studies suggest that the shorter isoform of HOXA9 might also play an important role in the cell transformation and leukemogenesis of MLL-rearranged leukemia through cooperating with the full-length HOXA9.
ACKNOWLEDGEMENTS
The authors thank Gregory Hannon, Scott Hammond and Lin He for providing retroviral vectors. This work was supported in part by the Leukemia & Lymphoma Society (LLS) Special Fellowship (Z.L.), Gabrielle's Angel Foundation for Cancer Research (Z.L.; J.C.; H.H.), the LLS Translational Research Grant (J.D.R. & J.C.), CALGB pilot grant 20801(J.D.R. & J.C.), National Institutes of Health (NIH) R01 grant CA127277 (J.C.), American Cancer Society (ACS) Research Scholar grant (J.C.), and the Fidelity Foundation (J.D.R. & J.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST The authors indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS Conception and design: Zejuan Li
Financial support: Zejuan Li, Hao Huang, Janet D. Rowley, Jianjun Chen
Collection and assembly of data: All authors
Data analysis and/or interpretation: Jianjun Chen and Zejuan Li
Experiments conduction: Miao He, Ping Chen, Stephen Arnovitz, Yuanyuan Li, Hao Huang, and Zejuan Li
Manuscript writing: Zejuan Li prepared the manuscript and all authors helped to revise the manuscript
Final approval of manuscript: All authors
REFERENCE
- [1].Barber BA, Rastegar M. Epigenetic control of Hox genes during neurogenesis, development, and disease. Ann Anat. 2010;192:261–74. doi: 10.1016/j.aanat.2010.07.009. [DOI] [PubMed] [Google Scholar]
- [2].Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–71. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- [3].Sitwala KV, Dandekar MN, Hess JL. HOX proteins and leukemia. Int J Clin Exp Pathol. 2008;1:461–74. [PMC free article] [PubMed] [Google Scholar]
- [4].Huang Y, Sitwala K, Bronstein J, et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119:388–98. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schnabel CA, Jacobs Y, Cleary ML. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19:608–16. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- [6].Hu YL, Fong S, Ferrell C, Largman C, Shen WF. HOXA9 modulates its oncogenic partner Meis1 to influence normal hematopoiesis. Mol Cell Biol. 2009;29:5181–92. doi: 10.1128/MCB.00545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–3. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lawrence HJ, Christensen J, Fong S, et al. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106:3988–94. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Eklund E. The role of hox proteins in leukemogenesis: insights into key regulatory events in hematopoiesis. Crit Rev Oncog. 2011;16:65–76. doi: 10.1615/critrevoncog.v16.i1-2.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Faber J, Krivtsov AV, Stubbs MC, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113:2375–85. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li Z, Huang H, Li Y, et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically-abnormal AML. Blood. 2012 doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li Z, Luo RT, Mi S, et al. Consistent deregulation of gene expression between human and murine MLL rearrangement leukemias. Cancer Res. 2009;69:1109–16. doi: 10.1158/0008-5472.CAN-08-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Andreeff M, Ruvolo V, Gadgil S, et al. HOX expression patterns identify a common signature for favorable AML. Leukemia. 2008;22:2041–7. doi: 10.1038/leu.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–7. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- [15].Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–16. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- [16].Kroon E, Krosl J, Thorsteinsdottir U, et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. Embo J. 1998;17:3714–25. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zeisig BB, Milne T, Garcia-Cuellar MP, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–28. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim MH, Chang HH, Shin C, et al. Genomic structure and sequence analysis of human HOXA-9. DNA Cell Biol. 1998;17:407–14. doi: 10.1089/dna.1998.17.407. [DOI] [PubMed] [Google Scholar]
- [19].Popovic R, Erfurth F, Zeleznik-Le N. Transcriptional complexity of the HOXA9 locus. Blood Cells Mol Dis. 2008;40:156–9. doi: 10.1016/j.bcmd.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dintilhac A, Bihan R, Guerrier D, Deschamps S, Pellerin I. A conserved non-homeodomain Hoxa9 isoform interacting with CBP is co-expressed with the 'typical' Hoxa9 protein during embryogenesis. Gene Expr Patterns. 2004;4:215–22. doi: 10.1016/j.modgep.2003.08.006. [DOI] [PubMed] [Google Scholar]
- [21].Fujimoto S, Araki K, Chisaka O, et al. Analysis of the murine Hoxa-9 cDNA: an alternatively spliced transcript encodes a truncated protein lacking the homeodomain. Gene. 1998;209:77–85. doi: 10.1016/s0378-1119(98)00014-6. [DOI] [PubMed] [Google Scholar]
- [22].He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Li Z, Lu J, Sun M, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105:15535–40. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mi S, Li Z, Chen P, et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc Natl Acad Sci U S A. 2010;107:3710–5. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bach C, Buhl S, Mueller D, et al. Leukemogenic transformation by HOXA cluster genes. Blood. 2010;115:2910–8. doi: 10.1182/blood-2009-04-216606. [DOI] [PubMed] [Google Scholar]
- [26].Calvo KR, Sykes DB, Pasillas M, Kamps MP. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced meis expression. Molecular and cellular biology. 2000;20:3274–85. doi: 10.1128/mcb.20.9.3274-3285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang Y, Morrone G, Zhang J, et al. CUL-4A stimulates ubiquitylation and degradation of the HOXA9 homeodomain protein. EMBO J. 2003;22:6057–67. doi: 10.1093/emboj/cdg577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shin J, Park B, Lee S, et al. A short isoform of human cytomegalovirus US3 functions as a dominant negative inhibitor of the full-length form. Journal of virology. 2006;80:5397–404. doi: 10.1128/JVI.02397-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li TW, Ting JH, Yokoyama NN, et al. Wnt activation and alternative promoter repression of LEF1 in colon cancer. Molecular and cellular biology. 2006;26:5284–99. doi: 10.1128/MCB.00105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harbor perspectives in biology. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Figtree GA, McDonald D, Watkins H, Channon KM. Truncated estrogen receptor alpha 46-kDa isoform in human endothelial cells: relationship to acute activation of nitric oxide synthase. Circulation. 2003;107:120–6. doi: 10.1161/01.cir.0000043805.11780.f5. [DOI] [PubMed] [Google Scholar]
- [32].Gough PJ, Greaves DR, Gordon S. A naturally occurring isoform of the human macrophage scavenger receptor (SR-A) gene generated by alternative splicing blocks modified LDL uptake. Journal of lipid research. 1998;39:531–43. [PubMed] [Google Scholar]
- [33].Oakley RH, Jewell CM, Yudt MR, Bofetiado DM, Cidlowski JA. The dominant negative activity of the human glucocorticoid receptor beta isoform. Specificity and mechanisms of action. The Journal of biological chemistry. 1999;274:27857–66. doi: 10.1074/jbc.274.39.27857. [DOI] [PubMed] [Google Scholar]
- [34].Zaika AI, Slade N, Erster SH, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. The Journal of experimental medicine. 2002;196:765–80. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chang CP, Shen WF, Rozenfeld S, et al. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995;9:663–74. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- [36].Thorsteinsdottir U, Kroon E, Jerome L, Blasi F, Sauvageau G. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol Cell Biol. 2001;21:224–34. doi: 10.1128/MCB.21.1.224-234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]