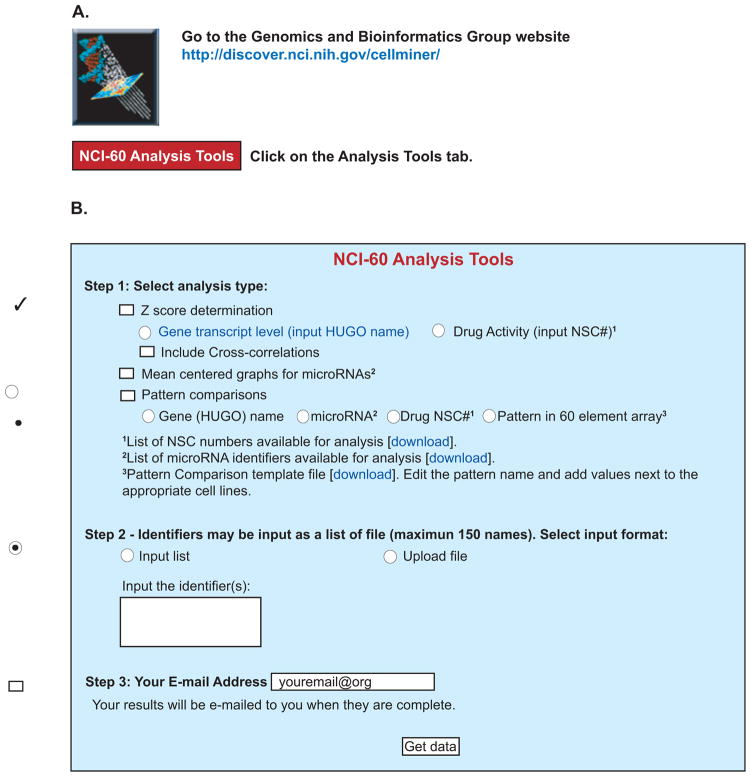
Figure 1.
Snapshot of the NCI-60 Analysis Website, a suite of web-based tools designed to facilitate rapid pharmacologic and genomic bioinformatics for the NCI-60 cell lines. A. These tools are accessible at the CellMiner website (http://discover.nci.nih.gov/cellminer/) by clicking on the NCI-60 Analysis Tools tab. B. The analysis of interest (Z score determination, Mean centered graphs for microRNAs, or Pattern comparisons) is selected in Step1 using the check boxes. The specific identifier or pattern of interest is selected in Step 2, either by typing in an identifier using the “Input list” function, or by uploading a file using the “Upload file” function. A maximum of 150 identifiers (genes, microRNAs or drugs) can be input at once. The results are e-mailed to the address entered in Step 3. Multiple check boxes may be selected for a single input. Radio buttons (circles) for an analysis type are mutually exclusive.