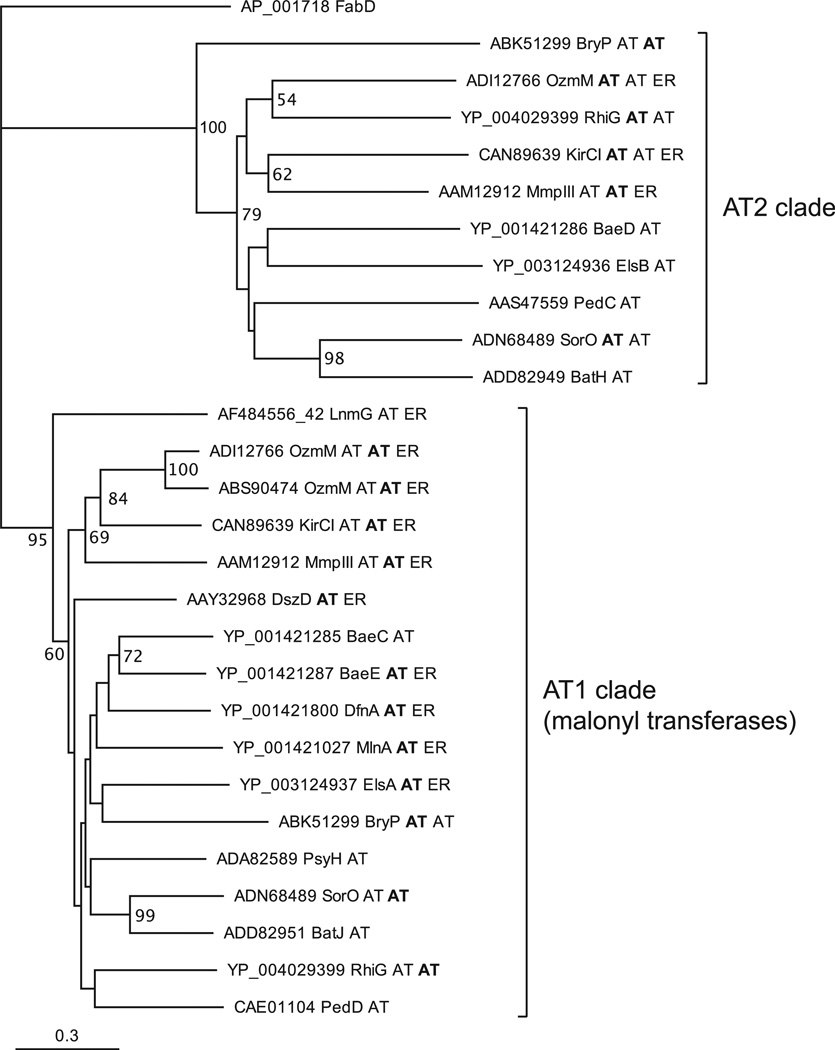
Figure 3. Phylogram of AT-like Enzymes from Various Trans-AT PKS Pathways.
Tip labels consist of accession numbers, protein name, and domain architecture. For multidomain proteins, the relevant domain is shown in bold. Sequences from the following pathways were used: Bae, bacillaene (B. amyloliquefaciens); Bat, batumin/kalimantacin (Pseudomonas fluorescens); Bry, bryostatin (“Candidatus Endobugula sertula”); Dfn, difficidin (B. amyloliquefaciens); Dsz, disorazol (Sorangium cellulosum); Els, elansolid (Chitinophaga pinensis); Kir, kirromycin (Streptomyces collinus); Lnm, leinamycin (Streptomyces atroolivaceus); Mmp, mupirocin (Pseudomonas fluorescens); Ozm, oxazolomycin (ADI12766 is from Streptomyces bingchenggensis and ABS90474 from Streptomyces albus); Psy, psymberin (uncultivated symbiont of sponge Psammocinia aff. bulbosa); Rhi, rhizoxin (Burkholderia rhizoxina); Sor, soraphen (Sorangium cellulosum). Outgroup is the E. coli AT from fatty acid biosynthesis (FabD).