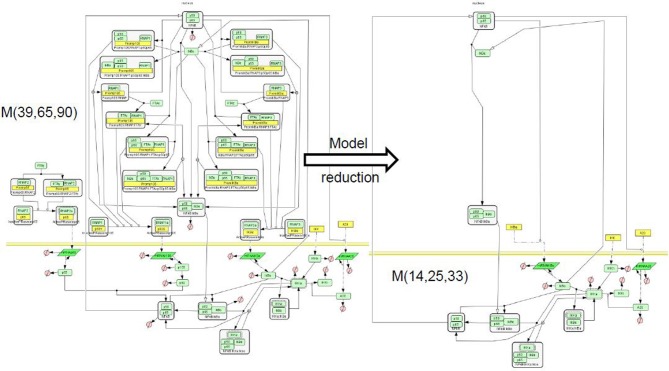
Figure 6.
Reduction of a NF-κB pathway model. We considered the model of NF-κB signaling [BIOMD0000000227 in Biomodels database (Le Novère et al., 2006), proposing separate production of the subunits p50, p65, the full combinatorics of their interactions as well as with the inhibitor IκB, the positive self-regulation of p50, and in addition an A20 molecule whose production is enhanced upon NF-κB stimulation, and which negatively regulates the activity of the stimulus responding kinase IKK (Radulescu et al., 2008). This model, denoted ℳ(39, 65, 90) contains 39 species, 65 reactions, and 90 parameters. We have reduced it to various levels of complexity. Among the reduced model we obtained one, ℳ(14, 25, 33) that has the same stoichiometry (but different rate functions) as a model published elsewhere by another author (Lipniacki et al., 2004) and denoted ℳ(14, 25, 28) (BIOMD0000000226 in Biomodels database). Incidently, this is also the simplest model in the hierarchy related to ℳ(39, 65, 90). Comparison (not shown) of the rate functions and of the trajectories of the models ℳ(14, 25, 33) and ℳ(14, 25, 28) provided insight into the consequences of various mechanistic modeling choices. The model graphical representation is based on the SBGN standard (Le Novère et al., 2009).