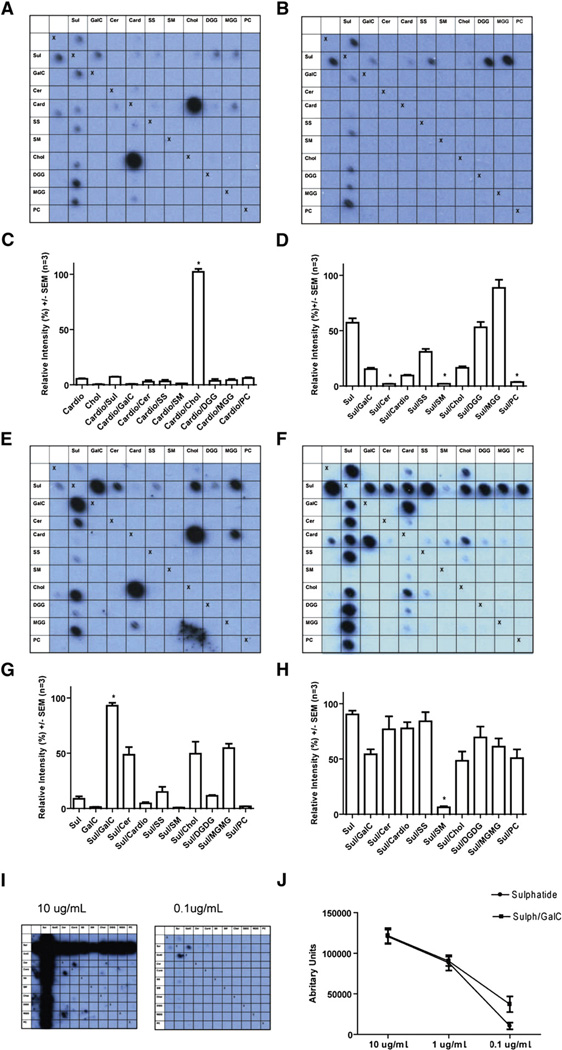
Fig. 2.
Illustrative blots and their quantitative analysis. Data were obtained using MS CSF (A–D) and recombinant antibodies 3, 73, 17 (Table 2) cloned from a single CSF-resident plasma cell (E–J). Row and column headings reveal the complex at each location. We used the following lipids either alone or in combination: sulfatide (Sul), galactocerebroside (GalC), ceramide (Cer), cardiolipin (Card), sphingosine (SS), sphingomyelin (SM), cholesterol (Chol), digalactosyl diglyceride (DGG), monogalactosyl diglyceride (MGG), phosphatidylcholine (PC). “Xs” represent the negative controls (methanol) which act as a line of symmetry for duplicate spots on the same membrane. (A, C) PPMS CSF blot and quantitative data, for binding to the complex formed by cholesterol and cardiolipin is increased by 96.3% compared to the sum of the intensities for the individual lipids (p = 0.0004, paired t test n=3). (B, D) Complex attenuation in a CSF MS blot, where binding to sulfatide yields a mean relative intensity signal of 57.1%, yet the intensity for the complexes of sulfatide created with the lipids ceramide, sphingomyelin and phosphatidylcholine is almost eliminated at between 1.96% and 3.66% (p<0.0001, GLM ANOVA with Tukey n = 3). (E, G) Blot and quantification from MS rAb 3 with anti-myelin lipid complex reactivity. Binding to the complex of sulfatide and galactocerebroside is increased by 82.84% (p = 0.004 paired t test, n = 3) compared to the sum of the mean intensities recorded from the individual lipids. (F, H) Blot and quantification from MS rAb 73 with binding to sulfatide that is inhibited when sulfatide is complexed with the lipid sphingomyelin (p<0.0001 GLM ANOVA with Tukey, n = 3). (I, J) MS rAb 17 assayed by serial dilution. The pattern of binding seen at higher rAb concentrations when signals are saturated appears largely complex independent. At a lower concentration of 0.1 µg/ml binding to the complex created by sulfatide and galactocerebroside can be seen to be enhanced (p = 0.0143 paired t test, n=3).