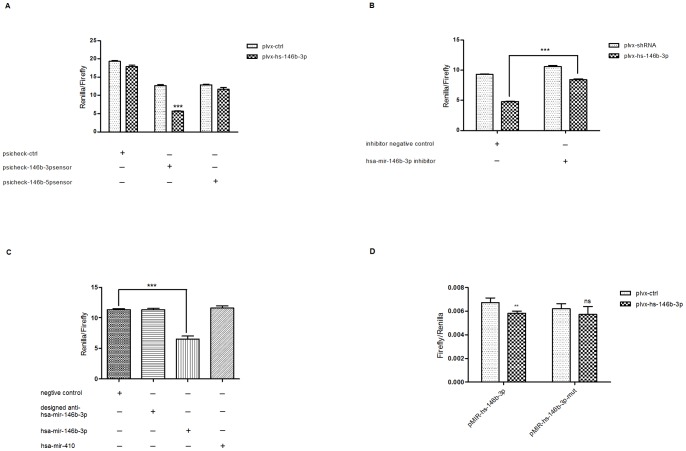
Figure 3. plvx-hs-146b-3p overexpressed functional hsa-miR-146b-3p with no detectable activity change of hsa-miR-146b-5p.
(A) Transcription products from plvx-hs-146b-3p had hsa-miR-146b-3p activities, without changing the activity of hsa-miR-146b-5p. HeLa cells were transfected with psicheck-ctrl (20 ng per well), psicheck-146b-3psensor (20 ng per well), or psicheck-146b-5psensor (20 ng per well), in combination with either plvx-ctrl (100 ng per well) or plvx-hs-146b-3p (100 ng per well), as indicated in the graph. (B) The repression of psicheck-146b-3psensor activity could be rescued using hsa-miR-146b-3p inhibitors. HeLa cells were transfected with psicheck-146b-3psensor (20 ng per well) in combination of plvx-ctrl (100 ng per well) or plvx-hs-146b-3p (100 ng per well) under the presentation of hsa-miR-146b-3p inhibitors (100 nM) or inhibitor negative controls (100 nM), as indicated in the graph. (C) Designed modified complementary sequences (designed anti-hsa-miR-146b-3p) could not repress psicheck-146b-3psensor activity as effectively as hsa-miR-146b-3p. HeLa cells were transfected with psicheck-146b-3psensor (20 ng per well) in combination with single stranded RNA mimics of negative control sequences (100 nM), designed anti-hsa-miR-146b-3p (100 nM), hsa-miR-146b-3p (100 nM), and hsa-miR-410 (100 nM), as indicated in the graph. The miRNA hsa-miR-410, which had no ability to repress psicheck-146b-3psensor activity, was used as a negative control miRNA. (D) The miRNA hsa-miR-146b-3p overexpressed by plvx-hs-146b-3p could repress the activity of bulged sensor of hsa-miR-146b-3p (pMIR-hs-146b-3p) and this repression was abolished by mutation of the seed sequences of the bulged sensor. HeLa cells were transfected with pMIR-hs-146b-3p (2 ng per well) or pMIR-hs-146b-3p-mut (2 ng per well), together with plvx-ctrl (400 ng per well) or plvx-hs-146b-3p (400 ng per well), as indicated in the graph. Renilla luciferase vector (5 ng per well) was delivered simultaneously as a transfection control. For all assays, protein was collected 24 h after transfection. Luciferase activity was quantified and expressed as relative luciferase activity. The data represent one of at least three independent experiments, each of which involved four replicates. Error bars represent SEMs. ** means p value ≤0.01; *** means p value ≤0.001; ns means no significance.